pbmc68k_reults
DongyueXie
2023-02-20
Last updated: 2023-02-20
Checks: 7 0
Knit directory: gsmash/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220606) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8044c17. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Untracked files:
Untracked: analysis/compare_vga_iterations_pbmc_full.Rmd
Untracked: output/pbmc3k_k1.rds
Untracked: output/pbmc3k_k1_S1.rds
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/overdispersed_splitting.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/pbmc68k_reults.Rmd) and
HTML (docs/pbmc68k_reults.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8044c17 | DongyueXie | 2023-02-20 | wflow_publish("analysis/pbmc68k_reults.Rmd") |
Introduction
Fresh 68k PBMCs (Donor A) Single Cell Gene Expression Dataset by Cell Ranger 1.1.0 Fresh 68k PBMCs (Donor A)
~68,000 cells detected
Sequenced on Illumina NextSeq 500 High Output with ~20,000 reads per cell
98bp read1 (transcript), 8bp I5 sample barcode, 14bp I7 GemCode barcode and 5bp read2 (UMI)
Analysis run with –cells=24000
I filtered out genes that expressed in less than 100 cells, and removed ‘CD34+’,‘CD4+ T Helper2’ cell types because there are less than 100 cells of each cell type. The final data matrix is
library(Matrix)
load("/project2/mstephens/pcarbo/git/single-cell-topics/data/pbmc_68k.RData")
counts = counts[,colSums(counts!=0)>100]
cell_idx = rowSums(counts!=0)>200 & !(samples$celltype %in%c('CD34+','CD4+ T Helper2'))
counts = counts[cell_idx,]
print(dim(counts))[1] 68186 11222Model fitting
The model is the Empirical Bayes Poisson matrix factorization model,
and \(l_0\) is fixed at
log(rowMeans(Y)), \(f_0\)
is initialized at log(colSums(Y)/sum(l_0)), then let the
model estimates \(f_0\) during
iterations.
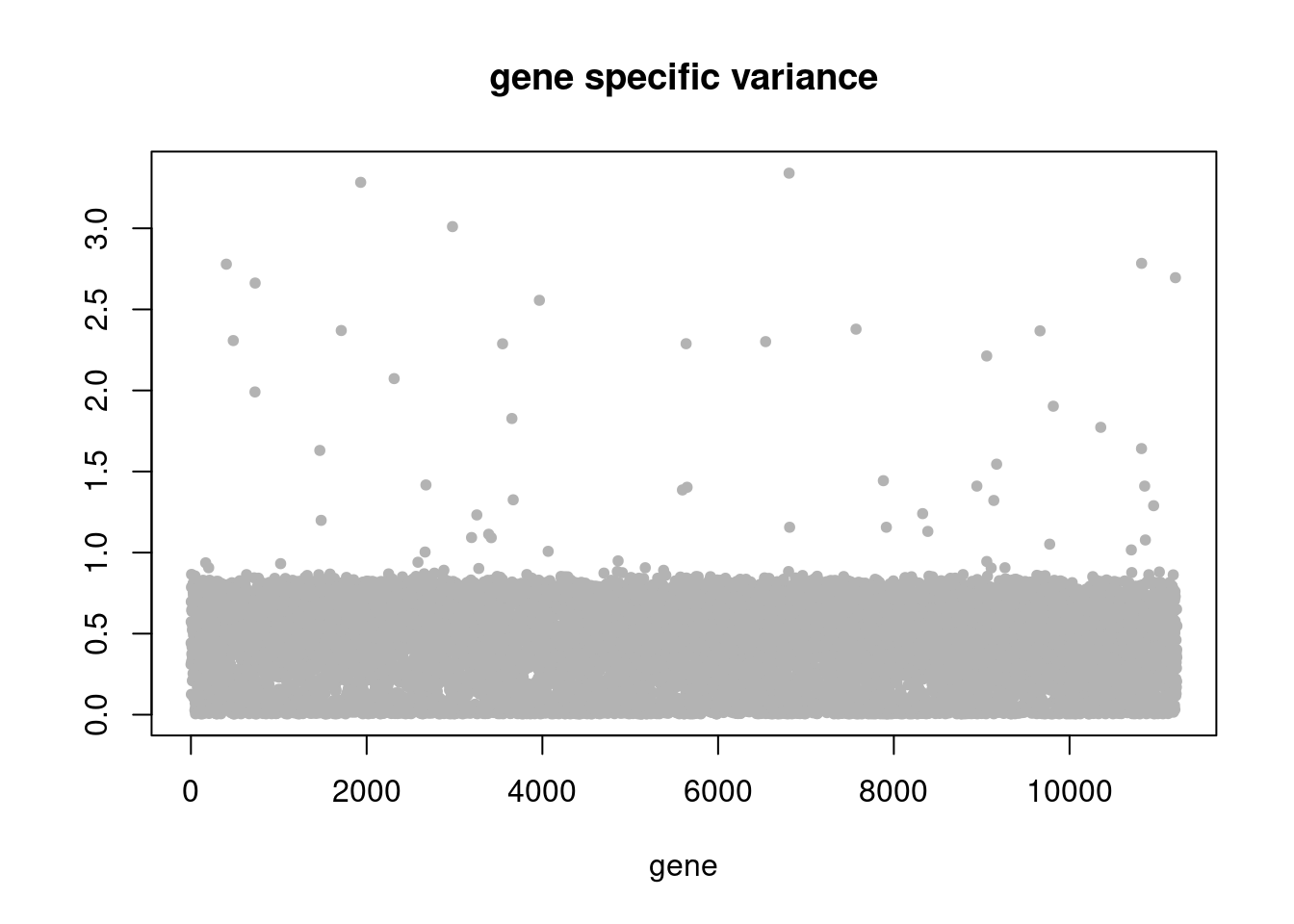
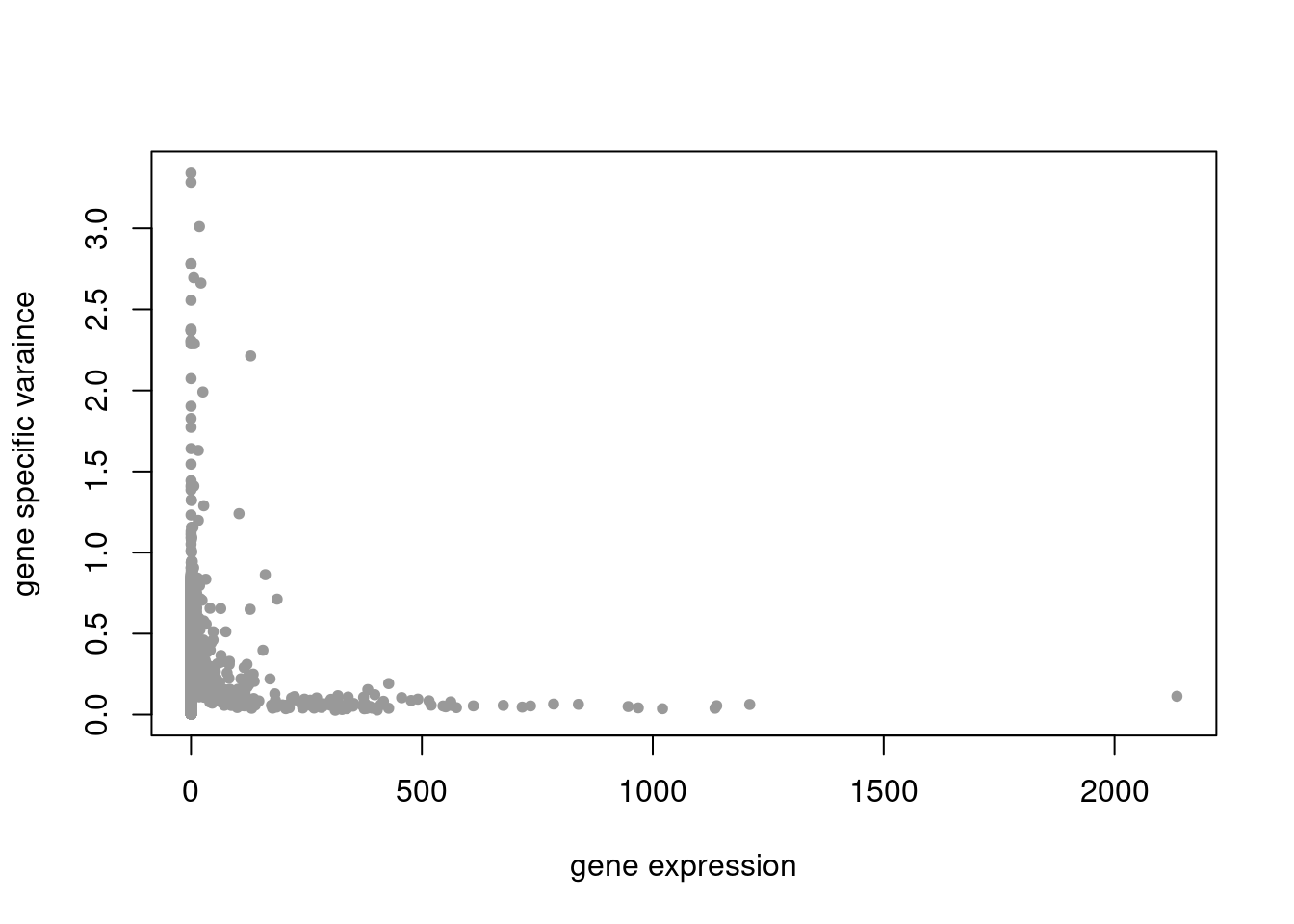
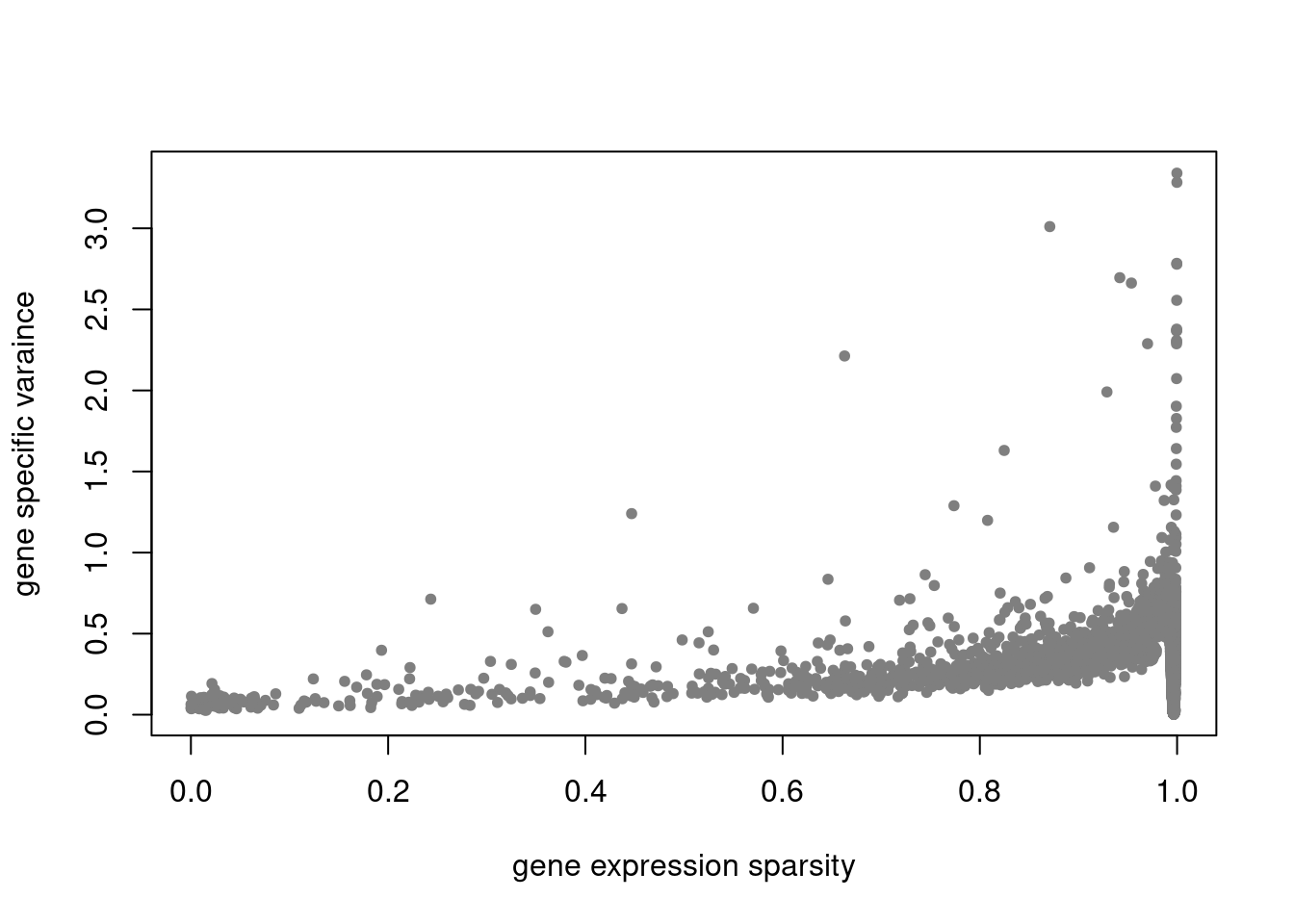
summary_plot = function(res,counts){
plot(res$K_trace,ylab='',xlab='iterations',main='K over iterations',pch=20)
plot(res$sigma2,ylab='',xlab='gene',main = 'gene specific variance',pch=20,col='grey70')
plot(colSums(counts/rowSums(counts)),res$sigma2,xlab='gene expression',ylab='gene specific varaince',pch=20,col='grey60')
plot(colSums(counts==0)/nrow(counts),res$sigma2,xlab='gene expression sparsity',ylab='gene specific varaince',pch = 20, col='grey50')
}
source('code/poisson_STM/plot_factors.R')
source('code/poisson_STM/structure_plot.R')Nonnegative loadings and factors with point exponential prior
res = readRDS('/project2/mstephens/dongyue/poisson_mf/pbmc_68k/ebpmf_pbmc_nonnegLF.rds')summary_plot(res,counts)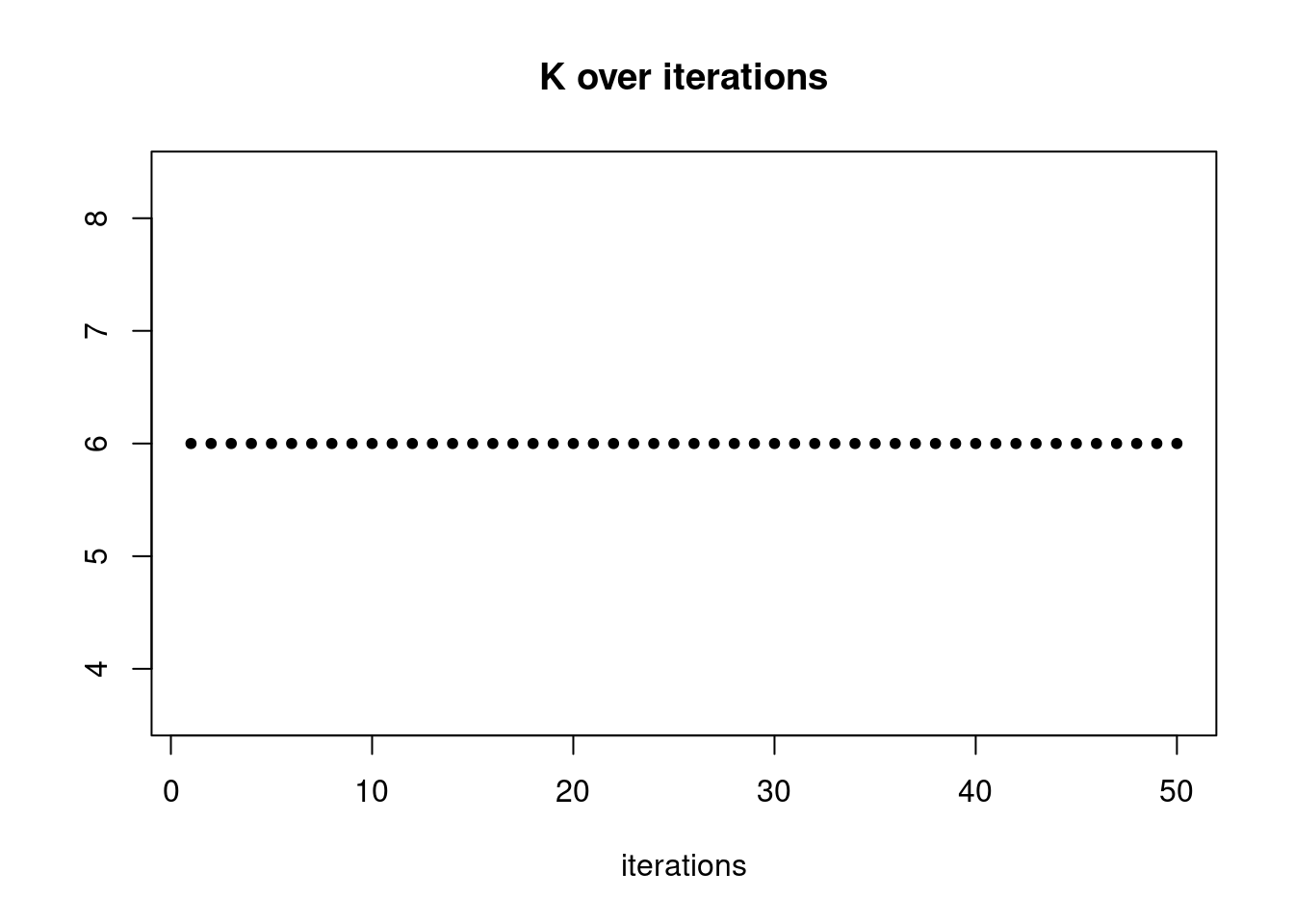
f0_init = log(colSums(counts)/sum(rowMeans(counts)))
plot(res$fit_flash$F.pm[,2],f0_init,xlab='f0 estimates',ylab='f0 init value',pch=20,col='grey70')
abline(a=0,b=1)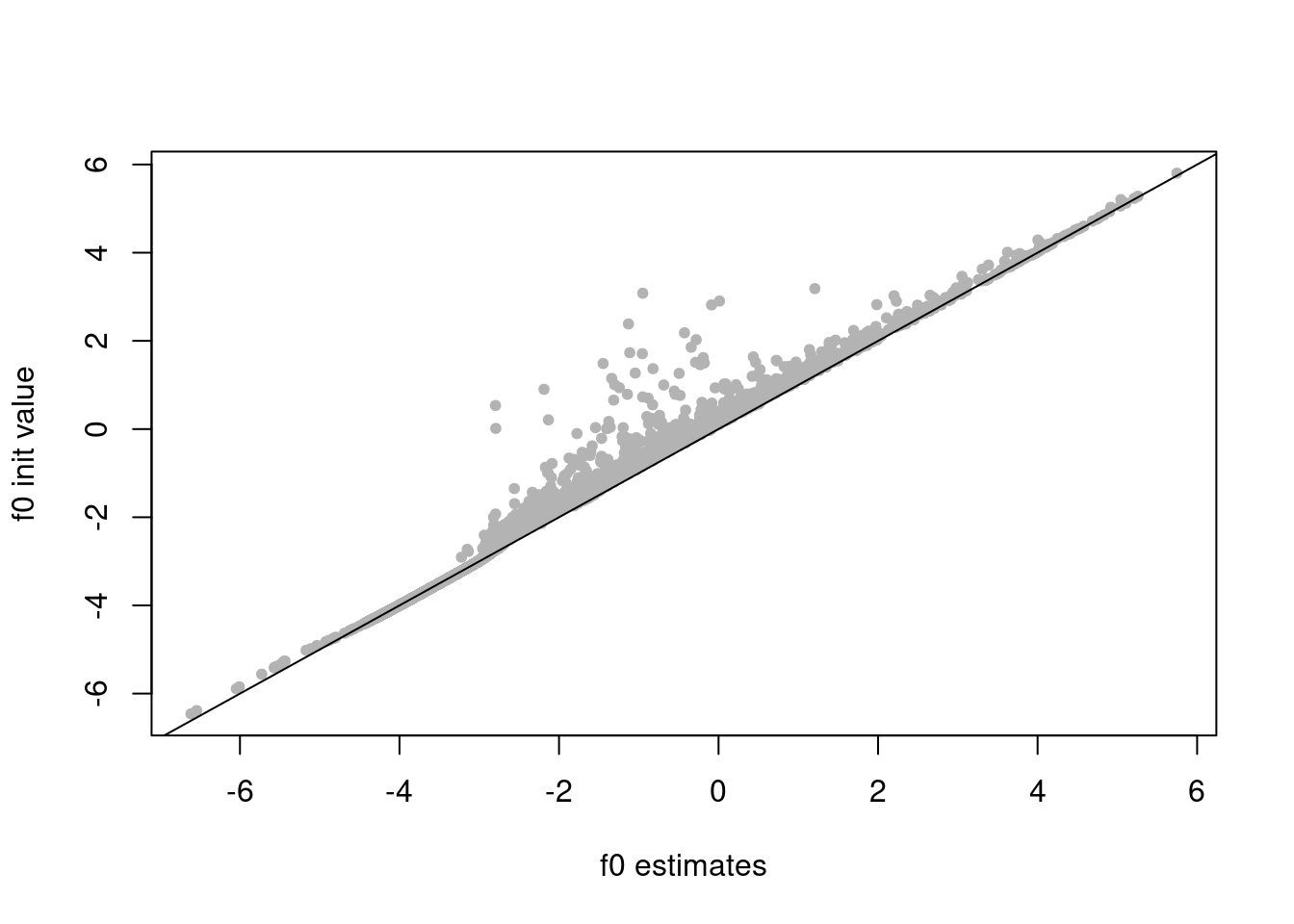
plot.factors(res$fit_flash,samples$celltype[cell_idx],nonnegative = T)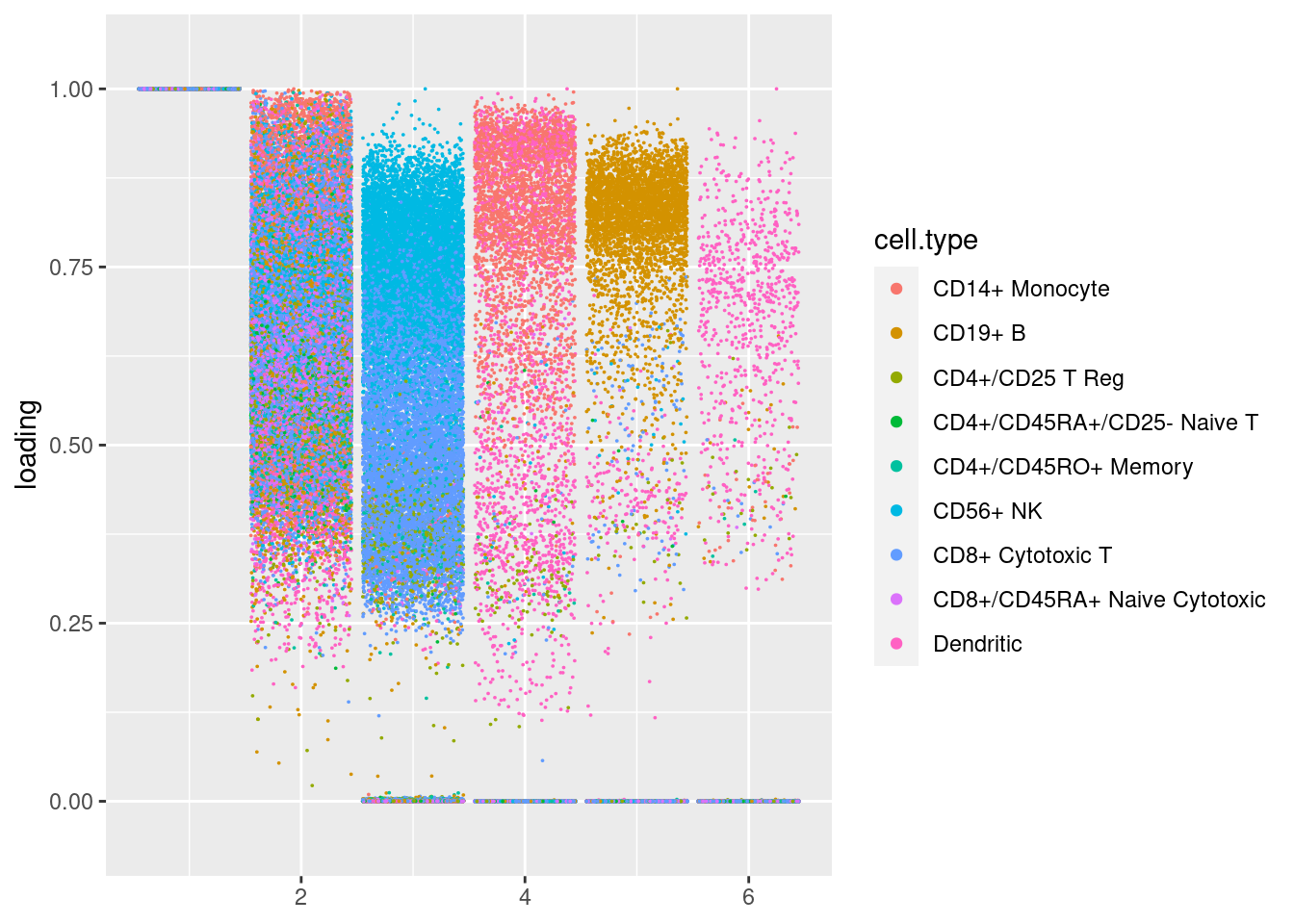
structure_plot_general(res$fit_flash$L.pm,res$fit_flash$F.pm,samples$celltype[cell_idx],LD=T)Running tsne on 90 x 4 matrix.Running tsne on 186 x 4 matrix.Running tsne on 174 x 4 matrix.Running tsne on 38 x 4 matrix.Running tsne on 86 x 4 matrix.Running tsne on 275 x 4 matrix.Running tsne on 599 x 4 matrix.Running tsne on 507 x 4 matrix.Running tsne on 45 x 4 matrix.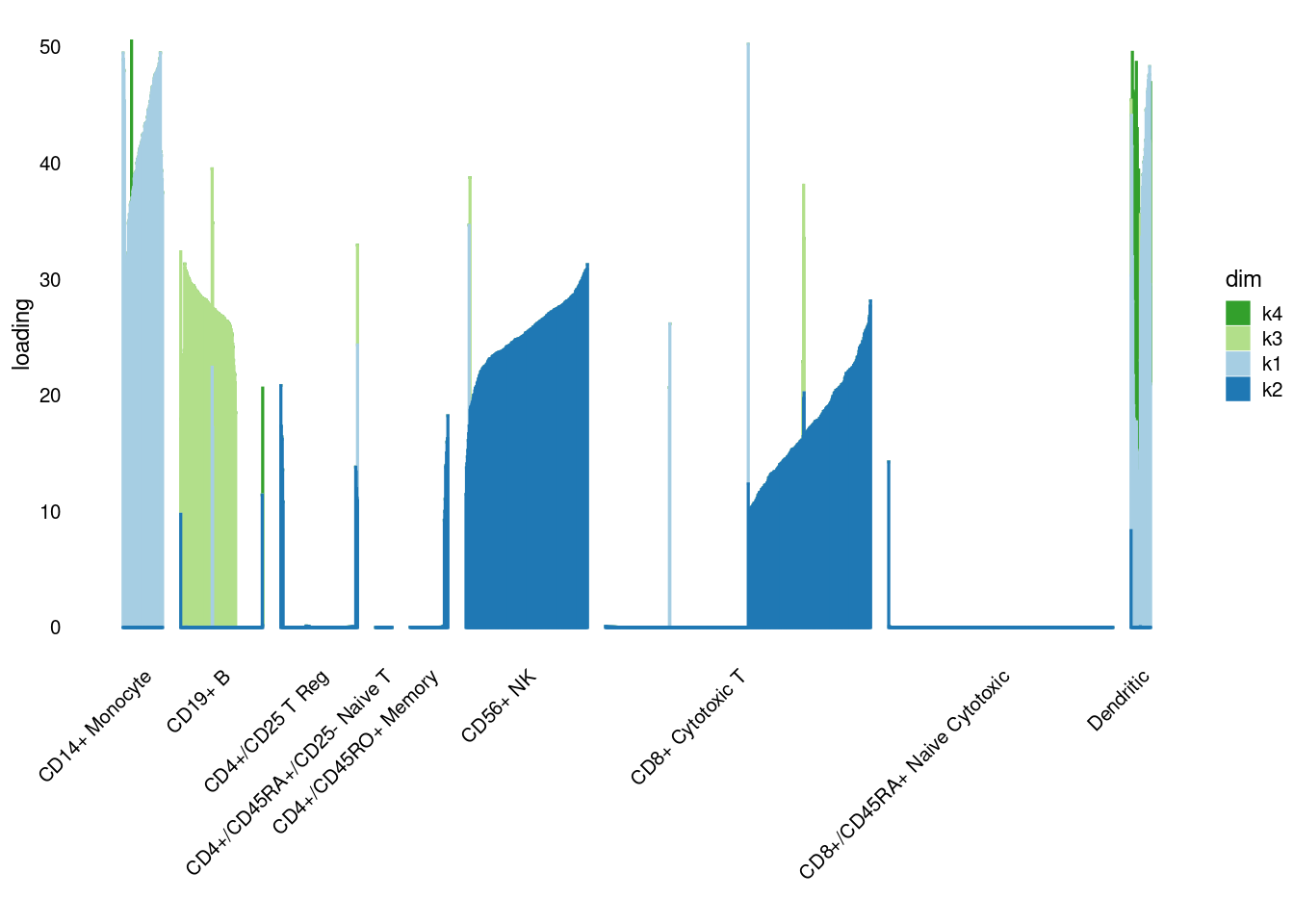
plot(res$fit_flash$pve[-c(1,2)],xlab='factors',ylab='PVE')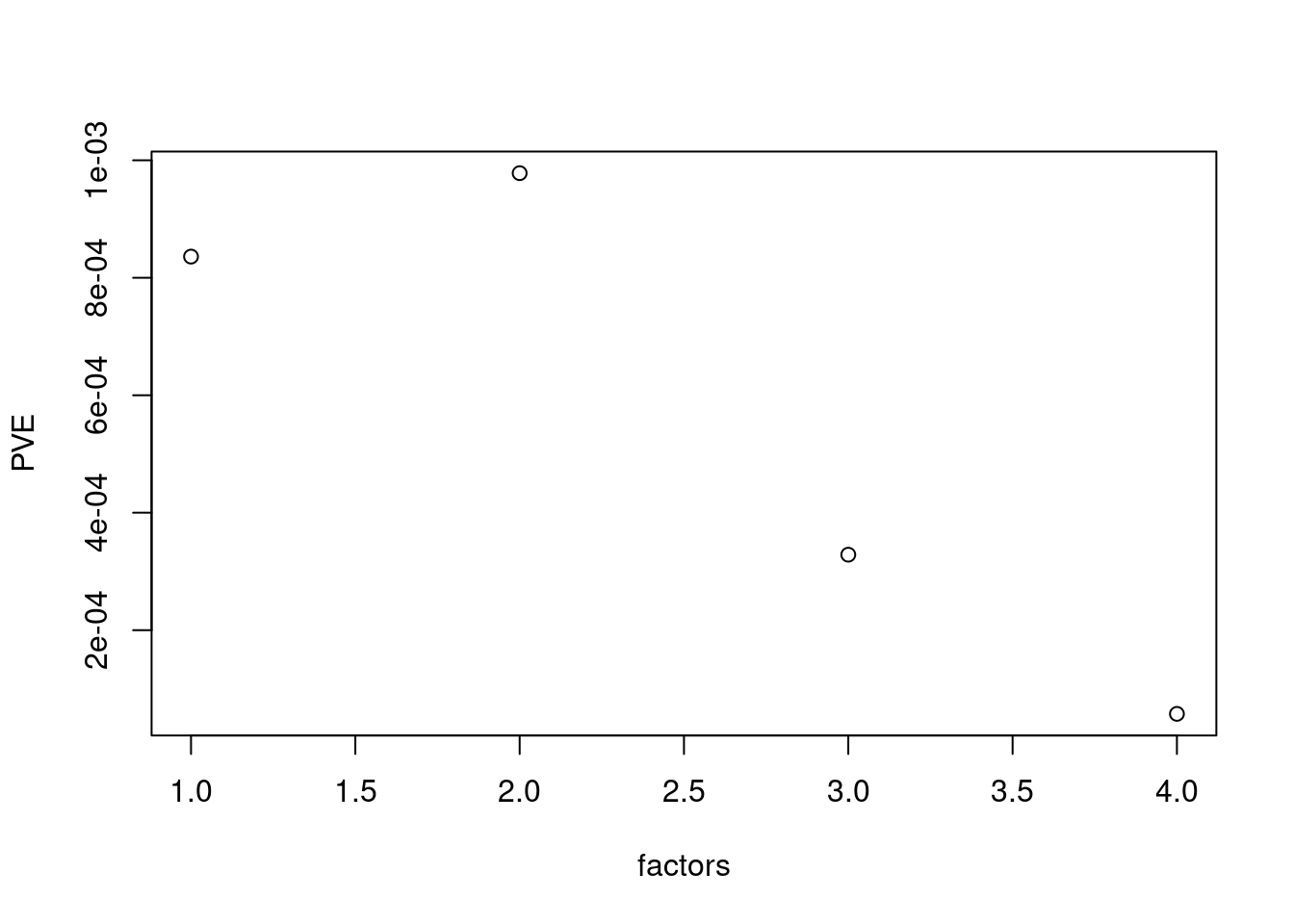
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /software/R-4.1.0-no-openblas-el7-x86_64/lib64/R/lib/libRblas.so
LAPACK: /software/R-4.1.0-no-openblas-el7-x86_64/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] fastTopics_0.6-142 ggplot2_3.4.0 Matrix_1.5-3 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] mcmc_0.9-7 bitops_1.0-7 matrixStats_0.59.0
[4] fs_1.5.0 progress_1.2.2 httr_1.4.4
[7] rprojroot_2.0.2 tools_4.1.0 bslib_0.2.5.1
[10] utf8_1.2.2 R6_2.5.1 irlba_2.3.5.1
[13] uwot_0.1.14 DBI_1.1.1 lazyeval_0.2.2
[16] colorspace_2.0-3 wavethresh_4.7.2 withr_2.5.0
[19] tidyselect_1.2.0 prettyunits_1.1.1 ebpm_0.0.1.3
[22] compiler_4.1.0 git2r_0.28.0 cli_3.5.0
[25] quantreg_5.94 SparseM_1.81 plotly_4.10.1
[28] labeling_0.4.2 horseshoe_0.2.0 sass_0.4.0
[31] smashrgen_1.1.4 caTools_1.18.2 flashier_0.2.34
[34] scales_1.2.1 SQUAREM_2021.1 quadprog_1.5-8
[37] pbapply_1.6-0 mixsqp_0.3-48 stringr_1.4.0
[40] digest_0.6.30 rmarkdown_2.9 MCMCpack_1.6-3
[43] deconvolveR_1.2-1 vebpm_0.4.3 pkgconfig_2.0.3
[46] htmltools_0.5.3 ebpmf_2.0.5 highr_0.9
[49] fastmap_1.1.0 invgamma_1.1 htmlwidgets_1.5.4
[52] rlang_1.0.6 rstudioapi_0.13 farver_2.1.1
[55] jquerylib_0.1.4 generics_0.1.3 jsonlite_1.8.3
[58] dplyr_1.0.10 magrittr_2.0.3 smashr_1.3-6
[61] Rcpp_1.0.9 munsell_0.5.0 fansi_1.0.3
[64] RcppZiggurat_0.1.6 lifecycle_1.0.3 stringi_1.6.2
[67] whisker_0.4 yaml_2.3.6 MASS_7.3-54
[70] plyr_1.8.6 Rtsne_0.16 grid_4.1.0
[73] parallel_4.1.0 promises_1.2.0.1 ggrepel_0.9.2
[76] crayon_1.5.2 lattice_0.20-44 cowplot_1.1.1
[79] splines_4.1.0 hms_1.1.2 knitr_1.33
[82] pillar_1.8.1 softImpute_1.4-1 reshape2_1.4.4
[85] glue_1.6.2 evaluate_0.14 trust_0.1-8
[88] data.table_1.14.6 RcppParallel_5.1.5 nloptr_1.2.2.2
[91] vctrs_0.5.1 httpuv_1.6.1 MatrixModels_0.5-1
[94] gtable_0.3.1 purrr_0.3.5 ebnm_1.0-11
[97] tidyr_1.2.1 assertthat_0.2.1 ashr_2.2-54
[100] xfun_0.24 Rfast_2.0.6 NNLM_0.4.4
[103] coda_0.19-4 later_1.3.0 survival_3.2-11
[106] viridisLite_0.4.1 truncnorm_1.0-8 tibble_3.1.8
[109] ellipsis_0.3.2