lasso_complexity
Matthew Stephens
2021-05-26
Last updated: 2021-05-26
Checks: 7 0
Knit directory: misc/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version edacee4. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/ALStruct_cache/
Ignored: data/.Rhistory
Ignored: data/pbmc/
Untracked files:
Untracked: .dropbox
Untracked: Icon
Untracked: analysis/GHstan.Rmd
Untracked: analysis/GTEX-cogaps.Rmd
Untracked: analysis/PACS.Rmd
Untracked: analysis/Rplot.png
Untracked: analysis/SPCAvRP.rmd
Untracked: analysis/admm_02.Rmd
Untracked: analysis/admm_03.Rmd
Untracked: analysis/compare-transformed-models.Rmd
Untracked: analysis/cormotif.Rmd
Untracked: analysis/cp_ash.Rmd
Untracked: analysis/eQTL.perm.rand.pdf
Untracked: analysis/eb_prepilot.Rmd
Untracked: analysis/eb_var.Rmd
Untracked: analysis/ebpmf1.Rmd
Untracked: analysis/flash_test_tree.Rmd
Untracked: analysis/flash_tree.Rmd
Untracked: analysis/ieQTL.perm.rand.pdf
Untracked: analysis/lasso_em_03.Rmd
Untracked: analysis/m6amash.Rmd
Untracked: analysis/mash_bhat_z.Rmd
Untracked: analysis/mash_ieqtl_permutations.Rmd
Untracked: analysis/mixsqp.Rmd
Untracked: analysis/mr.ash_lasso_init.Rmd
Untracked: analysis/mr.mash.test.Rmd
Untracked: analysis/mr_ash_modular.Rmd
Untracked: analysis/mr_ash_parameterization.Rmd
Untracked: analysis/mr_ash_ridge.Rmd
Untracked: analysis/mv_gaussian_message_passing.Rmd
Untracked: analysis/nejm.Rmd
Untracked: analysis/nmf_bg.Rmd
Untracked: analysis/normal_conditional_on_r2.Rmd
Untracked: analysis/normalize.Rmd
Untracked: analysis/pbmc.Rmd
Untracked: analysis/poisson_transform.Rmd
Untracked: analysis/pseudodata.Rmd
Untracked: analysis/qrnotes.txt
Untracked: analysis/ridge_iterative_02.Rmd
Untracked: analysis/ridge_iterative_splitting.Rmd
Untracked: analysis/samps/
Untracked: analysis/sc_bimodal.Rmd
Untracked: analysis/shrinkage_comparisons_changepoints.Rmd
Untracked: analysis/susie_en.Rmd
Untracked: analysis/susie_z_investigate.Rmd
Untracked: analysis/svd-timing.Rmd
Untracked: analysis/temp.RDS
Untracked: analysis/temp.Rmd
Untracked: analysis/test-figure/
Untracked: analysis/test.Rmd
Untracked: analysis/test.Rpres
Untracked: analysis/test.md
Untracked: analysis/test_qr.R
Untracked: analysis/test_sparse.Rmd
Untracked: analysis/z.txt
Untracked: code/multivariate_testfuncs.R
Untracked: code/rqb.hacked.R
Untracked: data/4matthew/
Untracked: data/4matthew2/
Untracked: data/E-MTAB-2805.processed.1/
Untracked: data/ENSG00000156738.Sim_Y2.RDS
Untracked: data/GDS5363_full.soft.gz
Untracked: data/GSE41265_allGenesTPM.txt
Untracked: data/Muscle_Skeletal.ACTN3.pm1Mb.RDS
Untracked: data/Thyroid.FMO2.pm1Mb.RDS
Untracked: data/bmass.HaemgenRBC2016.MAF01.Vs2.MergedDataSources.200kRanSubset.ChrBPMAFMarkerZScores.vs1.txt.gz
Untracked: data/bmass.HaemgenRBC2016.Vs2.NewSNPs.ZScores.hclust.vs1.txt
Untracked: data/bmass.HaemgenRBC2016.Vs2.PreviousSNPs.ZScores.hclust.vs1.txt
Untracked: data/eb_prepilot/
Untracked: data/finemap_data/fmo2.sim/b.txt
Untracked: data/finemap_data/fmo2.sim/dap_out.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2_snp.txt
Untracked: data/finemap_data/fmo2.sim/dap_out_snp.txt
Untracked: data/finemap_data/fmo2.sim/data
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.ld
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.z
Untracked: data/finemap_data/fmo2.sim/pos.txt
Untracked: data/logm.csv
Untracked: data/m.cd.RDS
Untracked: data/m.cdu.old.RDS
Untracked: data/m.new.cd.RDS
Untracked: data/m.old.cd.RDS
Untracked: data/mainbib.bib.old
Untracked: data/mat.csv
Untracked: data/mat.txt
Untracked: data/mat_new.csv
Untracked: data/matrix_lik.rds
Untracked: data/paintor_data/
Untracked: data/running_data_chris.csv
Untracked: data/running_data_matthew.csv
Untracked: data/temp.txt
Untracked: data/y.txt
Untracked: data/y_f.txt
Untracked: data/zscore_jointLCLs_m6AQTLs_susie_eQTLpruned.rds
Untracked: data/zscore_jointLCLs_random.rds
Untracked: explore_udi.R
Untracked: output/fit.k10.rds
Untracked: output/fit.varbvs.RDS
Untracked: output/glmnet.fit.RDS
Untracked: output/test.bv.txt
Untracked: output/test.gamma.txt
Untracked: output/test.hyp.txt
Untracked: output/test.log.txt
Untracked: output/test.param.txt
Untracked: output/test2.bv.txt
Untracked: output/test2.gamma.txt
Untracked: output/test2.hyp.txt
Untracked: output/test2.log.txt
Untracked: output/test2.param.txt
Untracked: output/test3.bv.txt
Untracked: output/test3.gamma.txt
Untracked: output/test3.hyp.txt
Untracked: output/test3.log.txt
Untracked: output/test3.param.txt
Untracked: output/test4.bv.txt
Untracked: output/test4.gamma.txt
Untracked: output/test4.hyp.txt
Untracked: output/test4.log.txt
Untracked: output/test4.param.txt
Untracked: output/test5.bv.txt
Untracked: output/test5.gamma.txt
Untracked: output/test5.hyp.txt
Untracked: output/test5.log.txt
Untracked: output/test5.param.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/lasso_complexity.Rmd) and HTML (docs/lasso_complexity.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | edacee4 | Matthew Stephens | 2021-05-26 | workflowr::wflow_publish(“lasso_complexity.Rmd”) |
| html | 84f23cb | Matthew Stephens | 2021-05-26 | Build site. |
| Rmd | 840b89e | Matthew Stephens | 2021-05-26 | wflow_publish(“lasso_complexity.Rmd”) |
| html | 222582b | Matthew Stephens | 2021-05-26 | Build site. |
| Rmd | b4fb979 | Matthew Stephens | 2021-05-26 | wflow_publish(“lasso_complexity.Rmd”) |
Introduction
I was suprised to hear that (“worst case”) lasso complexity for full solution path is O(np min(n,p)). I always thought it was O(np) per iteration and did not think about how the number of iterations required might increase with n and p. I wanted to do a quick simulation check.
Empirically the log-log plot for time vs n is approximately linear with slope near 1.25, suggesting a practical scaling of O(n^1.25 p). Of course this is very much just a quick initial assessment.
set.seed(123)
library("glmnet")Warning: package 'glmnet' was built under R version 3.6.2Loading required package: MatrixLoaded glmnet 4.1n_seq = c(100, 200, 500, 1000, 2000, 5000, 10000)
p = 10000
nmax = 10000
X = matrix(rnorm(nmax*p),nrow=nmax)
b = rnorm(p)
time = c()
for(n in n_seq){
y = X[1:n,] %*% b + rnorm(n)
time = c(time,system.time(fit <- glmnet(X[1:n,],y))[1]) # user time
#print(time)
}
plot(log(n_seq),log(time), main = "log(user time) vs log(n)")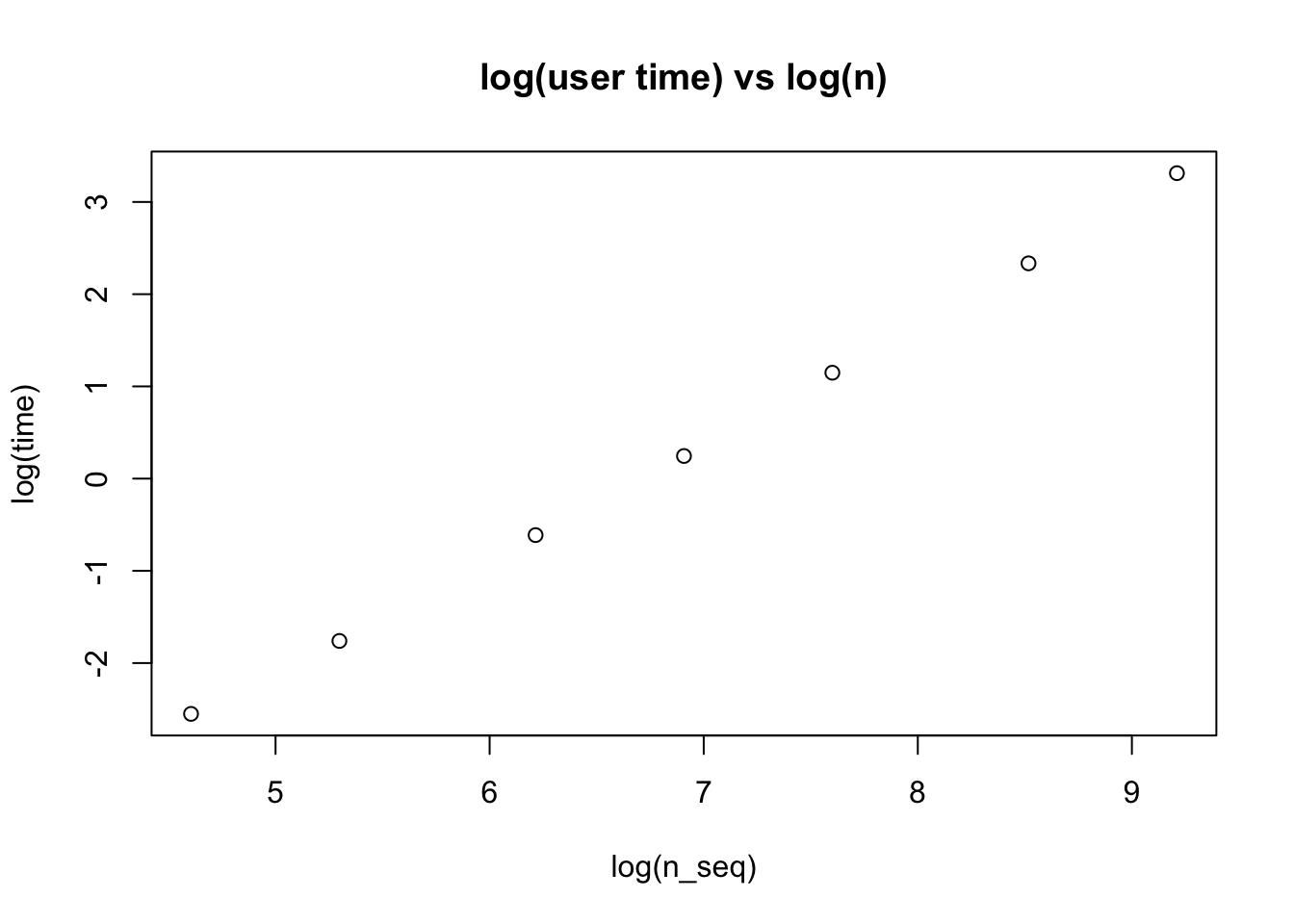
slope = (log(time)[7]-log(time)[1])/(log(n_seq)[7]-log(n_seq)[1])
print(slope)user.self
1.2732 Comparison with Susie
Just for interest I ran susie on the same datasets, but it is a bit slow for “dense” scenarios like this so I reduced p to 5k to save some time.
set.seed(123)
library("susieR")
n_seq = c(100, 200, 500, 1000, 2000, 5000)
p = 5000
nmax = max(n_seq)
X = matrix(rnorm(nmax*p),nrow=nmax)
b = rnorm(p)
time = c()
time.susie = c()
for(n in n_seq){
y = X[1:n,] %*% b + rnorm(n)
time.susie = c(time.susie,system.time(fit <- susie(X[1:n,],y))[1]) # user time
time = c(time, system.time(fit <- glmnet(X[1:n,],y))[1])
#print(time.susie)
}
plot(log(n_seq),log(time.susie), main = "red=susie; black=lasso",col=2,ylim=c(-3.5,4))
points(log(n_seq),log(time))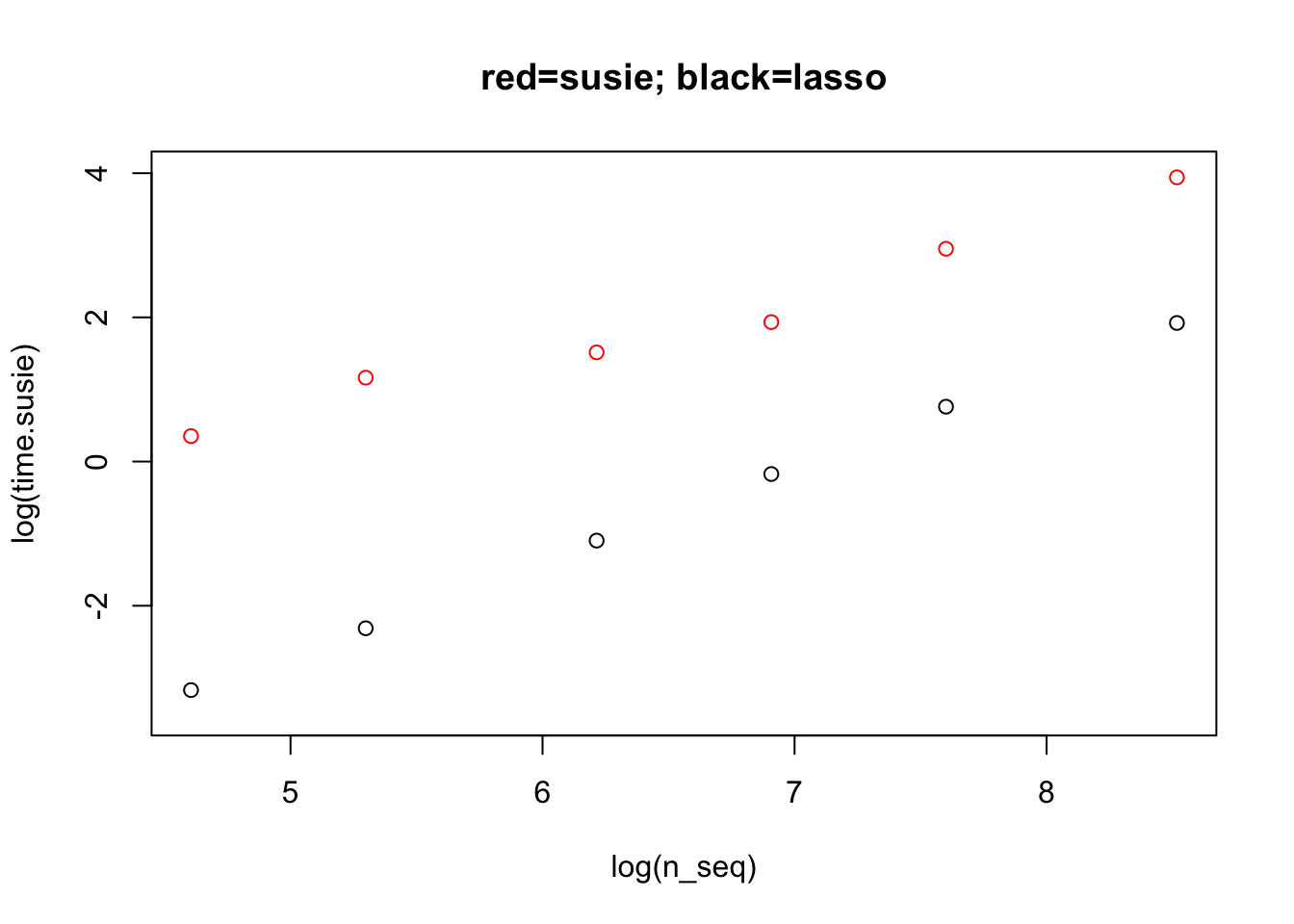
For the largest dataset here susie is slower than a single fit of lasso (complete solution path) by a factor of 7.5335772. This is without CV for lasso though, so with 5-fold or 10-fold CV the times would be comparable.
Sparser case
Here I simulate data with only 5 non-zero effects to see how it changes things. Here for the largest data-sets the two running times are similar (so susie would be faster if we did 5-fold CV for lasso).
set.seed(123)
n_seq = c(100, 200, 500, 1000, 2000, 5000)
p = 5000
nmax = max(n_seq)
X = matrix(rnorm(nmax*p),nrow=nmax)
b = rep(0,p)
b[1:5] = rnorm(5)
time = c()
time.susie = c()
for(n in n_seq){
y = X[1:n,] %*% b + rnorm(n)
time.susie = c(time.susie,system.time(fit <- susie(X[1:n,],y))[1]) # user time
time = c(time, system.time(fit <- glmnet(X[1:n,],y))[1])
#print(time.susie)
}
plot(log(n_seq),log(time.susie), main = "red=susie; black=lasso",col=2,ylim=c(-3.5,4))
points(log(n_seq),log(time))
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] susieR_0.11.26 glmnet_4.1 Matrix_1.2-18
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 plyr_1.8.6 pillar_1.4.6 compiler_3.6.0
[5] later_1.1.0.1 git2r_0.27.1 workflowr_1.6.2 iterators_1.0.12
[9] tools_3.6.0 digest_0.6.27 gtable_0.3.0 evaluate_0.14
[13] lifecycle_1.0.0 tibble_3.0.4 lattice_0.20-41 pkgconfig_2.0.3
[17] rlang_0.4.10 foreach_1.5.0 rstudioapi_0.13 yaml_2.2.1
[21] xfun_0.16 dplyr_1.0.2 stringr_1.4.0 knitr_1.29
[25] generics_0.0.2 fs_1.5.0 vctrs_0.3.8 tidyselect_1.1.0
[29] rprojroot_1.3-2 grid_3.6.0 reshape_0.8.8 glue_1.4.2
[33] R6_2.4.1 survival_3.2-3 rmarkdown_2.3 mixsqp_0.3-43
[37] irlba_2.3.3 purrr_0.3.4 ggplot2_3.3.2 magrittr_2.0.1
[41] whisker_0.4 scales_1.1.1 backports_1.1.10 promises_1.1.1
[45] codetools_0.2-16 ellipsis_0.3.1 htmltools_0.5.0 splines_3.6.0
[49] colorspace_1.4-1 shape_1.4.4 httpuv_1.5.4 stringi_1.4.6
[53] munsell_0.5.0 crayon_1.3.4