mr.ash.changepoint
Matthew Stephens
2019-11-01
Last updated: 2019-11-02
Checks: 7 0
Knit directory: misc/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/ALStruct_cache/
Ignored: data/.Rhistory
Ignored: data/pbmc/
Ignored: docs/figure/.DS_Store
Untracked files:
Untracked: .dropbox
Untracked: Icon
Untracked: analysis/GTEX-cogaps.Rmd
Untracked: analysis/PACS.Rmd
Untracked: analysis/SPCAvRP.rmd
Untracked: analysis/compare-transformed-models.Rmd
Untracked: analysis/cormotif.Rmd
Untracked: analysis/cp_ash.Rmd
Untracked: analysis/eQTL.perm.rand.pdf
Untracked: analysis/eb_prepilot.Rmd
Untracked: analysis/flash_test_tree.Rmd
Untracked: analysis/ieQTL.perm.rand.pdf
Untracked: analysis/m6amash.Rmd
Untracked: analysis/mash_bhat_z.Rmd
Untracked: analysis/mash_ieqtl_permutations.Rmd
Untracked: analysis/mixsqp.Rmd
Untracked: analysis/mr_ash_modular.Rmd
Untracked: analysis/mr_ash_parameterization.Rmd
Untracked: analysis/nejm.Rmd
Untracked: analysis/normalize.Rmd
Untracked: analysis/pbmc.Rmd
Untracked: analysis/poisson_transform.Rmd
Untracked: analysis/pseudodata.Rmd
Untracked: analysis/ridge_iterative_splitting.Rmd
Untracked: analysis/sc_bimodal.Rmd
Untracked: analysis/susie_en.Rmd
Untracked: analysis/susie_z_investigate.Rmd
Untracked: analysis/svd-timing.Rmd
Untracked: analysis/temp.Rmd
Untracked: analysis/test-figure/
Untracked: analysis/test.Rmd
Untracked: analysis/test.Rpres
Untracked: analysis/test.md
Untracked: analysis/test_sparse.Rmd
Untracked: analysis/z.txt
Untracked: code/multivariate_testfuncs.R
Untracked: data/4matthew/
Untracked: data/4matthew2/
Untracked: data/E-MTAB-2805.processed.1/
Untracked: data/ENSG00000156738.Sim_Y2.RDS
Untracked: data/GDS5363_full.soft.gz
Untracked: data/GSE41265_allGenesTPM.txt
Untracked: data/Muscle_Skeletal.ACTN3.pm1Mb.RDS
Untracked: data/Thyroid.FMO2.pm1Mb.RDS
Untracked: data/bmass.HaemgenRBC2016.MAF01.Vs2.MergedDataSources.200kRanSubset.ChrBPMAFMarkerZScores.vs1.txt.gz
Untracked: data/bmass.HaemgenRBC2016.Vs2.NewSNPs.ZScores.hclust.vs1.txt
Untracked: data/bmass.HaemgenRBC2016.Vs2.PreviousSNPs.ZScores.hclust.vs1.txt
Untracked: data/eb_prepilot/
Untracked: data/finemap_data/fmo2.sim/b.txt
Untracked: data/finemap_data/fmo2.sim/dap_out.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2_snp.txt
Untracked: data/finemap_data/fmo2.sim/dap_out_snp.txt
Untracked: data/finemap_data/fmo2.sim/data
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.ld
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.z
Untracked: data/finemap_data/fmo2.sim/pos.txt
Untracked: data/logm.csv
Untracked: data/m.cd.RDS
Untracked: data/m.cdu.old.RDS
Untracked: data/m.new.cd.RDS
Untracked: data/m.old.cd.RDS
Untracked: data/mainbib.bib.old
Untracked: data/mat.csv
Untracked: data/mat.txt
Untracked: data/mat_new.csv
Untracked: data/matrix_lik.rds
Untracked: data/paintor_data/
Untracked: data/temp.txt
Untracked: data/y.txt
Untracked: data/y_f.txt
Untracked: data/zscore_jointLCLs_m6AQTLs_susie_eQTLpruned.rds
Untracked: data/zscore_jointLCLs_random.rds
Untracked: docs/figure/cp_ash.Rmd/
Untracked: docs/figure/eigen.Rmd/
Untracked: docs/figure/fmo2.sim.Rmd/
Untracked: docs/figure/mr_ash_modular.Rmd/
Untracked: docs/figure/newVB.elbo.Rmd/
Untracked: docs/figure/poisson_transform.Rmd/
Untracked: docs/figure/rbc_zscore_mash2.Rmd/
Untracked: docs/figure/rbc_zscore_mash2_analysis.Rmd/
Untracked: docs/figure/rbc_zscores.Rmd/
Untracked: docs/figure/ridge_iterative_splitting.Rmd/
Untracked: docs/figure/susie_en.Rmd/
Untracked: docs/figure/test.Rmd/
Untracked: docs/trend_files/
Untracked: docs/z.txt
Untracked: explore_udi.R
Untracked: output/fit.k10.rds
Untracked: output/fit.varbvs.RDS
Untracked: output/glmnet.fit.RDS
Untracked: output/test.bv.txt
Untracked: output/test.gamma.txt
Untracked: output/test.hyp.txt
Untracked: output/test.log.txt
Untracked: output/test.param.txt
Untracked: output/test2.bv.txt
Untracked: output/test2.gamma.txt
Untracked: output/test2.hyp.txt
Untracked: output/test2.log.txt
Untracked: output/test2.param.txt
Untracked: output/test3.bv.txt
Untracked: output/test3.gamma.txt
Untracked: output/test3.hyp.txt
Untracked: output/test3.log.txt
Untracked: output/test3.param.txt
Untracked: output/test4.bv.txt
Untracked: output/test4.gamma.txt
Untracked: output/test4.hyp.txt
Untracked: output/test4.log.txt
Untracked: output/test4.param.txt
Untracked: output/test5.bv.txt
Untracked: output/test5.gamma.txt
Untracked: output/test5.hyp.txt
Untracked: output/test5.log.txt
Untracked: output/test5.param.txt
Unstaged changes:
Modified: analysis/minque.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0e9e766 | Matthew Stephens | 2019-11-02 | workflowr::wflow_publish(“mr.ash.changepoint.Rmd”) |
| html | 15c0630 | Matthew Stephens | 2019-11-02 | Build site. |
| Rmd | a0bc392 | Matthew Stephens | 2019-11-02 | workflowr::wflow_publish(“mr.ash.changepoint.Rmd”) |
library("mr.ash.alpha")
library("glmnet")Loading required package: MatrixLoading required package: foreachLoaded glmnet 2.0-18library("genlasso")Loading required package: igraph
Attaching package: 'igraph'The following objects are masked from 'package:stats':
decompose, spectrumThe following object is masked from 'package:base':
unionIntroduction
I’m going to try the current version of mr.ash.alpha on a changepoint problem for my own interest.
Single changepoint
First simulate data:
set.seed(100)
n = 100
p = n
X = matrix(0,nrow=n,ncol=n)
for(i in 1:n){
X[i:n,i] = 1
}
btrue = rep(0,n)
btrue[50] = 8
Y = X %*% btrue + rnorm(n)
plot(Y)
lines(X %*% btrue)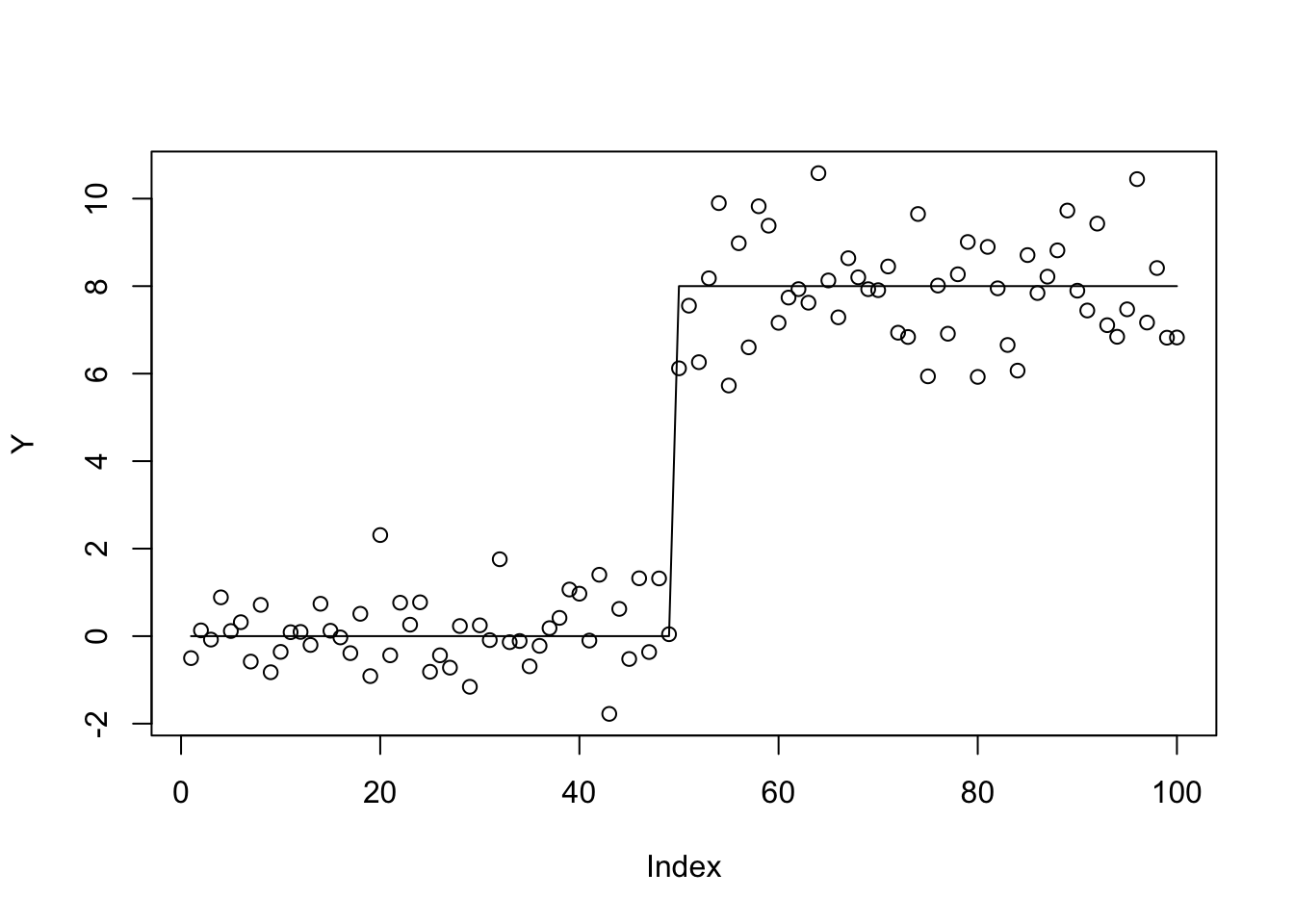
| Version | Author | Date |
|---|---|---|
| 15c0630 | Matthew Stephens | 2019-11-02 |
This works great out of the box:
fit.ma = mr.ash(X,Y)
plot(Y)
lines(X %*% btrue,lwd=1)
lines(predict(fit.ma,X),col=2,lwd=2)
fit.glmnet = glmnet(X,Y)
fit.glmnet.cv = glmnet::cv.glmnet(X,Y)
lines(predict(fit.glmnet.cv,X),col=3,lwd=2)
fit.genlasso = trendfilter(Y,ord=0)
fit.genlasso.cv = cv.trendfilter(fit.genlasso)Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... lines(fit.genlasso$fit[,which(fit.genlasso.cv$lambda==fit.genlasso.cv$lambda.min)],lwd=2,col=4)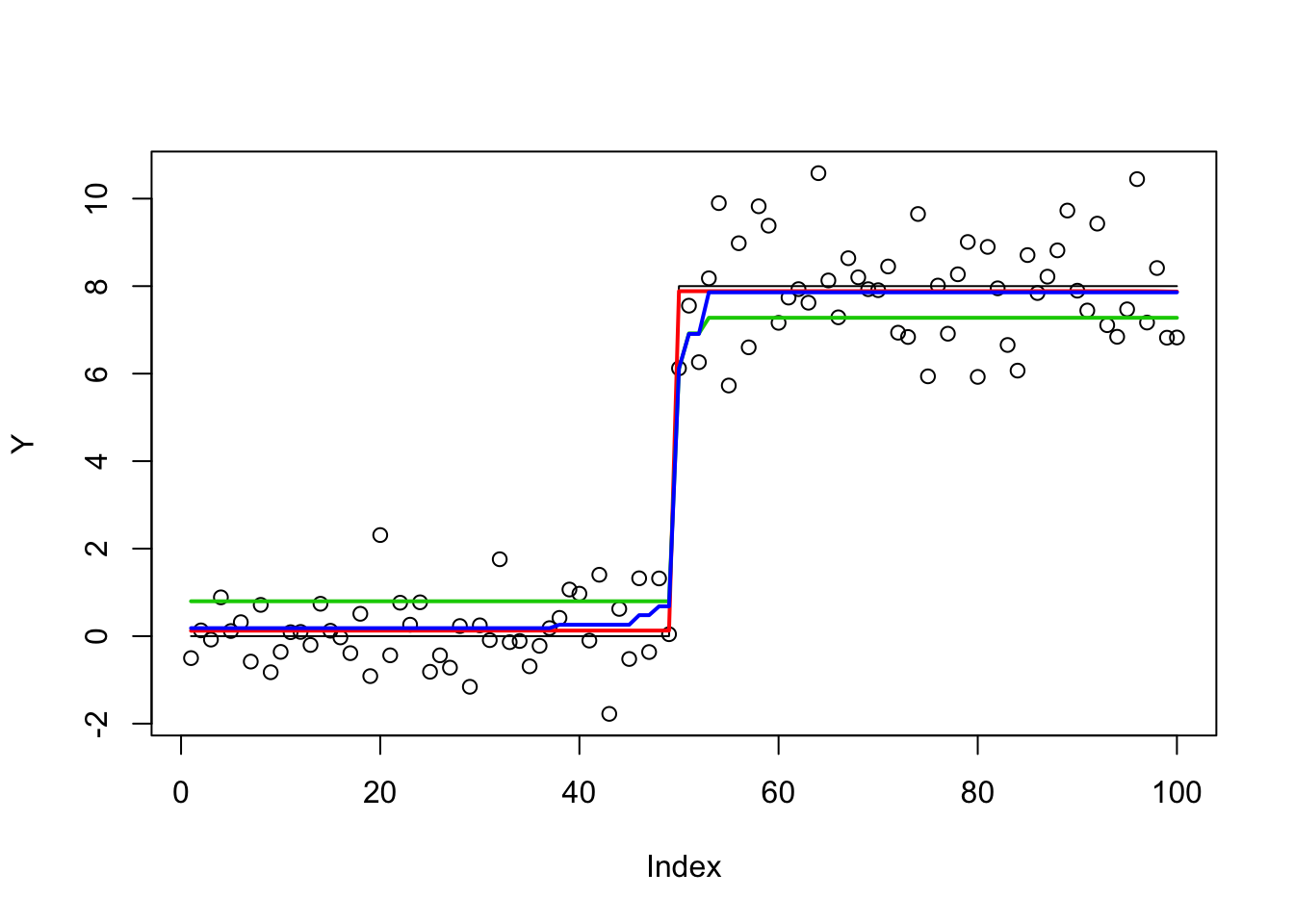
| Version | Author | Date |
|---|---|---|
| 15c0630 | Matthew Stephens | 2019-11-02 |
Double changepoint
Now we do a harder case, similar to the Susie paper:
btrue[52] = -8
Y = X %*% btrue + rnorm(n,0,0.1)
plot(Y)
lines(X %*% btrue)
| Version | Author | Date |
|---|---|---|
| 15c0630 | Matthew Stephens | 2019-11-02 |
fit.ma = mr.ash(X,Y)
plot(Y)
lines(predict(fit.ma,X),col=2)
fit.glmnet = glmnet(X,Y)
fit.glmnet.cv = glmnet::cv.glmnet(X,Y)
lines(predict(fit.glmnet.cv,X),col=3,lwd=2)
fit.genlasso = trendfilter(Y,ord=0)
fit.genlasso.cv = cv.trendfilter(fit.genlasso)Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... lines(fit.genlasso$fit[,which(fit.genlasso.cv$lambda==fit.genlasso.cv$lambda.min)],lwd=2,col=4)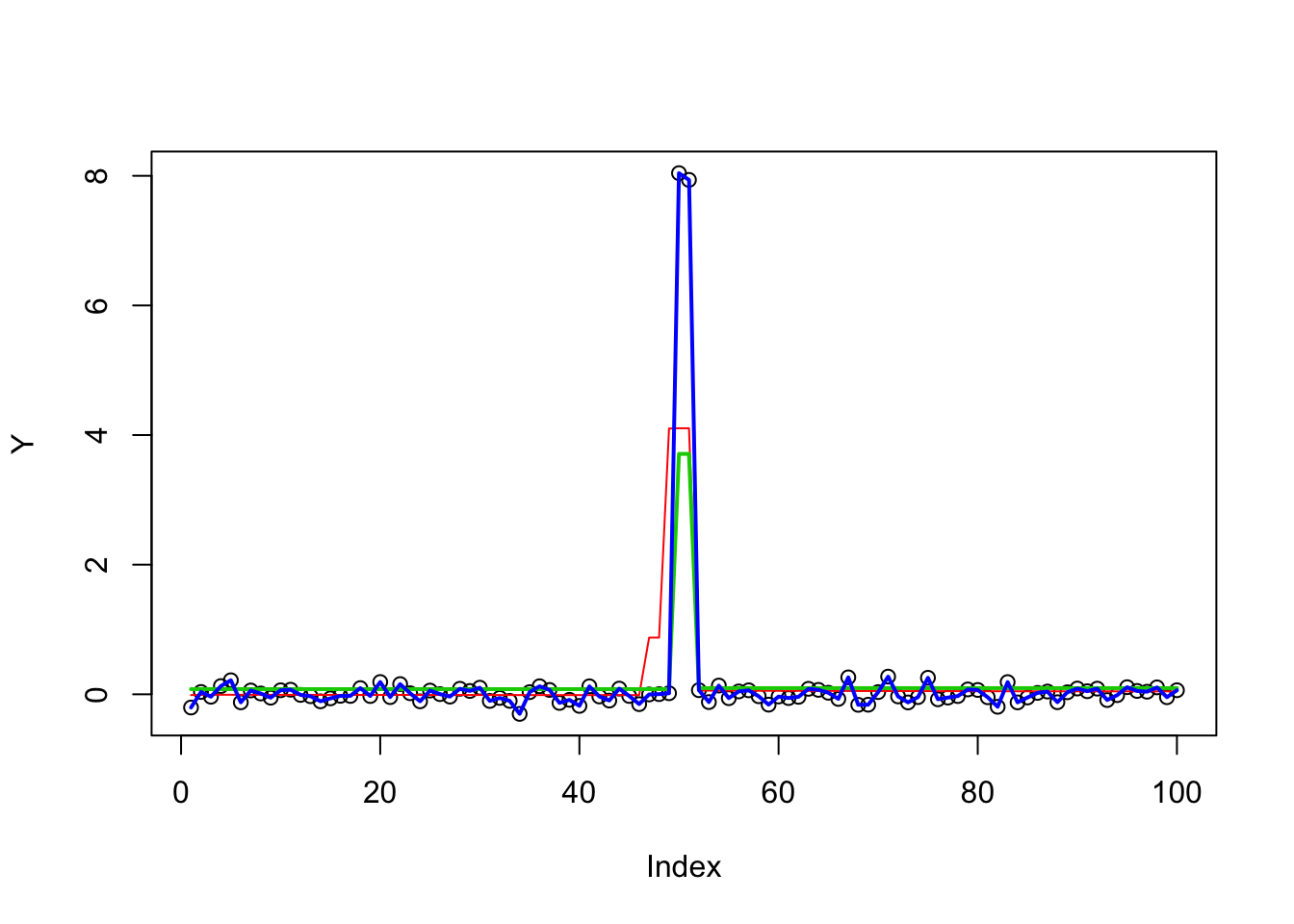
| Version | Author | Date |
|---|---|---|
| 15c0630 | Matthew Stephens | 2019-11-02 |
It is interesting that glmnet and genlasso give quite different answers. Clearly glmnet is entirely missing the changepoint. If you zoom in you will see that genlasso is overfitting - it is not just getting the single changepoint but fitting the data everywhere!
Also it looks like in this case mr.ash is converging to local optima. (the change is a bit too early…) Here we try warmstarts of mr.ash from genlasso solution, and from truth. However, both give NaN as answers. Not sure why.
b.genlasso = fit.genlasso$beta[,which(fit.genlasso.cv$lambda==fit.genlasso.cv$lambda.min)]
plot(Y)
lines(b.genlasso)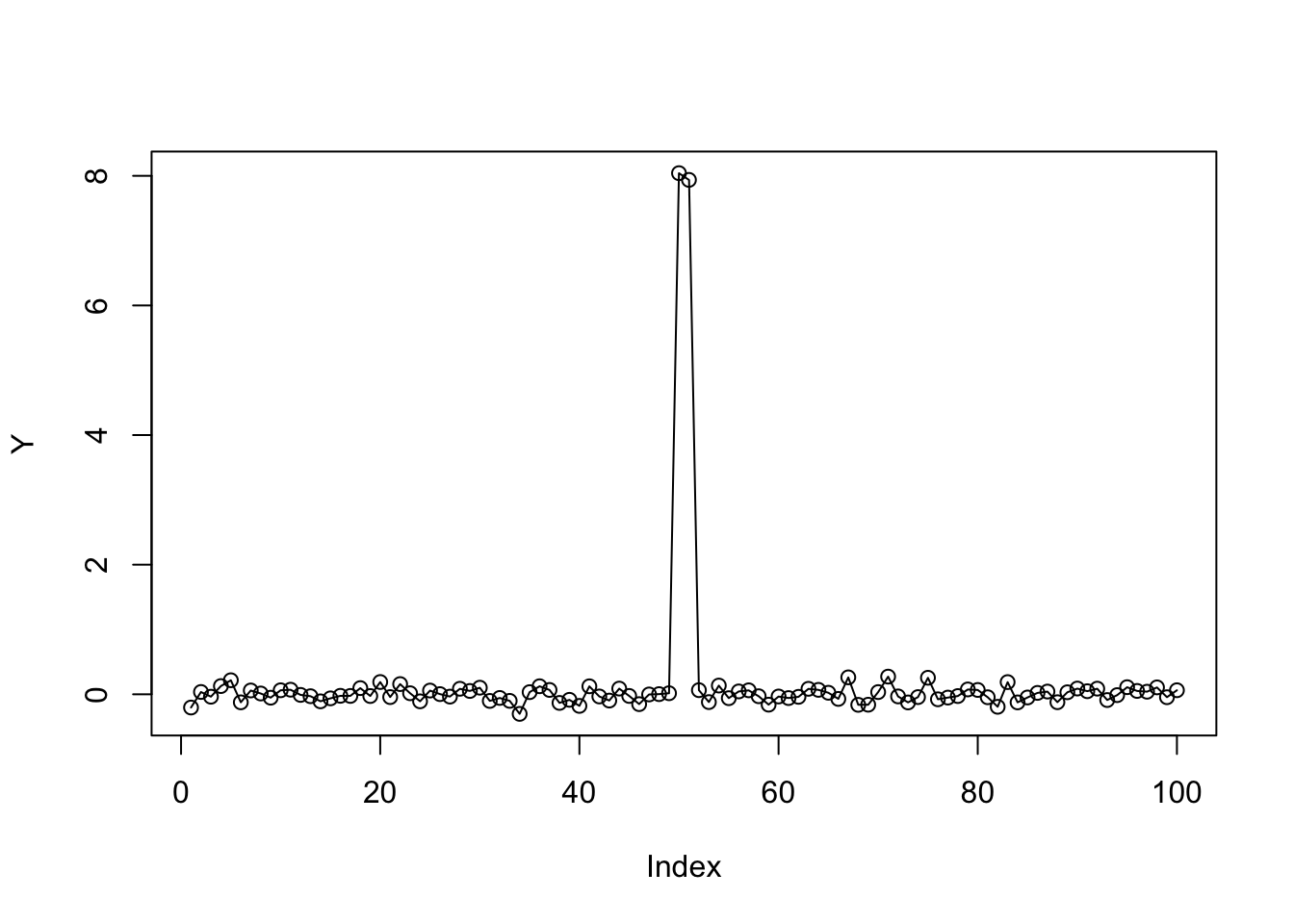
| Version | Author | Date |
|---|---|---|
| 15c0630 | Matthew Stephens | 2019-11-02 |
fit.ma.warm = mr.ash(X,Y,beta.init = b.genlasso)
fit.ma.warm2 = mr.ash(X,Y,beta.init = btrue)
predict(fit.ma.warm,X) [1] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[18] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[35] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[52] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[69] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[86] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaNpredict(fit.ma.warm2,X) [1] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[18] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[35] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[52] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[69] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
[86] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] genlasso_1.4 igraph_1.2.4.1 glmnet_2.0-18
[4] foreach_1.4.7 Matrix_1.2-17 mr.ash.alpha_0.1-3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.2 knitr_1.23 whisker_0.3-2 magrittr_1.5
[5] workflowr_1.4.0 lattice_0.20-38 stringr_1.4.0 tools_3.6.0
[9] grid_3.6.0 xfun_0.8 git2r_0.26.1 htmltools_0.3.6
[13] iterators_1.0.12 yaml_2.2.0 rprojroot_1.3-2 digest_0.6.20
[17] fs_1.3.1 codetools_0.2-16 glue_1.3.1 evaluate_0.14
[21] rmarkdown_1.14 stringi_1.4.3 compiler_3.6.0 backports_1.1.4
[25] pkgconfig_2.0.2