power_nneg
Matthew Stephens
2025-03-03
Last updated: 2025-03-09
Checks: 7 0
Knit directory: misc/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the
code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5d10e53. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/ALStruct_cache/
Ignored: data/.Rhistory
Ignored: data/methylation-data-for-matthew.rds
Ignored: data/pbmc/
Ignored: data/pbmc_purified.RData
Untracked files:
Untracked: .dropbox
Untracked: Icon
Untracked: analysis/GHstan.Rmd
Untracked: analysis/GTEX-cogaps.Rmd
Untracked: analysis/PACS.Rmd
Untracked: analysis/Rplot.png
Untracked: analysis/SPCAvRP.rmd
Untracked: analysis/abf_comparisons.Rmd
Untracked: analysis/admm_02.Rmd
Untracked: analysis/admm_03.Rmd
Untracked: analysis/bispca.Rmd
Untracked: analysis/cache/
Untracked: analysis/cholesky.Rmd
Untracked: analysis/compare-transformed-models.Rmd
Untracked: analysis/cormotif.Rmd
Untracked: analysis/cp_ash.Rmd
Untracked: analysis/eQTL.perm.rand.pdf
Untracked: analysis/eb_prepilot.Rmd
Untracked: analysis/eb_var.Rmd
Untracked: analysis/ebpmf1.Rmd
Untracked: analysis/ebpmf_sla_text.Rmd
Untracked: analysis/ebpower.Rmd
Untracked: analysis/ebspca_sims.Rmd
Untracked: analysis/explore_psvd.Rmd
Untracked: analysis/fa_check_identify.Rmd
Untracked: analysis/fa_iterative.Rmd
Untracked: analysis/flash_cov_overlapping_groups_init.Rmd
Untracked: analysis/flash_test_tree.Rmd
Untracked: analysis/flashier_newgroups.Rmd
Untracked: analysis/flashier_nmf_triples.Rmd
Untracked: analysis/flashier_pbmc.Rmd
Untracked: analysis/flashier_snn_shifted_prior.Rmd
Untracked: analysis/greedy_ebpmf_exploration_00.Rmd
Untracked: analysis/ieQTL.perm.rand.pdf
Untracked: analysis/lasso_em_03.Rmd
Untracked: analysis/m6amash.Rmd
Untracked: analysis/mash_bhat_z.Rmd
Untracked: analysis/mash_ieqtl_permutations.Rmd
Untracked: analysis/methylation_example.Rmd
Untracked: analysis/mixsqp.Rmd
Untracked: analysis/mr.ash_lasso_init.Rmd
Untracked: analysis/mr.mash.test.Rmd
Untracked: analysis/mr_ash_modular.Rmd
Untracked: analysis/mr_ash_parameterization.Rmd
Untracked: analysis/mr_ash_ridge.Rmd
Untracked: analysis/mv_gaussian_message_passing.Rmd
Untracked: analysis/nejm.Rmd
Untracked: analysis/nmf_bg.Rmd
Untracked: analysis/nonneg_underapprox.Rmd
Untracked: analysis/normal_conditional_on_r2.Rmd
Untracked: analysis/normalize.Rmd
Untracked: analysis/pbmc.Rmd
Untracked: analysis/pca_binary_weighted.Rmd
Untracked: analysis/pca_l1.Rmd
Untracked: analysis/poisson_nmf_approx.Rmd
Untracked: analysis/poisson_shrink.Rmd
Untracked: analysis/poisson_transform.Rmd
Untracked: analysis/qrnotes.txt
Untracked: analysis/ridge_iterative_02.Rmd
Untracked: analysis/ridge_iterative_splitting.Rmd
Untracked: analysis/samps/
Untracked: analysis/sc_bimodal.Rmd
Untracked: analysis/shrinkage_comparisons_changepoints.Rmd
Untracked: analysis/susie_cov.Rmd
Untracked: analysis/susie_en.Rmd
Untracked: analysis/susie_z_investigate.Rmd
Untracked: analysis/svd-timing.Rmd
Untracked: analysis/temp.RDS
Untracked: analysis/temp.Rmd
Untracked: analysis/test-figure/
Untracked: analysis/test.Rmd
Untracked: analysis/test.Rpres
Untracked: analysis/test.md
Untracked: analysis/test_qr.R
Untracked: analysis/test_sparse.Rmd
Untracked: analysis/tree_dist_top_eigenvector.Rmd
Untracked: analysis/z.txt
Untracked: code/multivariate_testfuncs.R
Untracked: code/rqb.hacked.R
Untracked: data/4matthew/
Untracked: data/4matthew2/
Untracked: data/E-MTAB-2805.processed.1/
Untracked: data/ENSG00000156738.Sim_Y2.RDS
Untracked: data/GDS5363_full.soft.gz
Untracked: data/GSE41265_allGenesTPM.txt
Untracked: data/Muscle_Skeletal.ACTN3.pm1Mb.RDS
Untracked: data/P.rds
Untracked: data/Thyroid.FMO2.pm1Mb.RDS
Untracked: data/bmass.HaemgenRBC2016.MAF01.Vs2.MergedDataSources.200kRanSubset.ChrBPMAFMarkerZScores.vs1.txt.gz
Untracked: data/bmass.HaemgenRBC2016.Vs2.NewSNPs.ZScores.hclust.vs1.txt
Untracked: data/bmass.HaemgenRBC2016.Vs2.PreviousSNPs.ZScores.hclust.vs1.txt
Untracked: data/eb_prepilot/
Untracked: data/finemap_data/fmo2.sim/b.txt
Untracked: data/finemap_data/fmo2.sim/dap_out.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2_snp.txt
Untracked: data/finemap_data/fmo2.sim/dap_out_snp.txt
Untracked: data/finemap_data/fmo2.sim/data
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.ld
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.z
Untracked: data/finemap_data/fmo2.sim/pos.txt
Untracked: data/logm.csv
Untracked: data/m.cd.RDS
Untracked: data/m.cdu.old.RDS
Untracked: data/m.new.cd.RDS
Untracked: data/m.old.cd.RDS
Untracked: data/mainbib.bib.old
Untracked: data/mat.csv
Untracked: data/mat.txt
Untracked: data/mat_new.csv
Untracked: data/matrix_lik.rds
Untracked: data/paintor_data/
Untracked: data/running_data_chris.csv
Untracked: data/running_data_matthew.csv
Untracked: data/temp.txt
Untracked: data/y.txt
Untracked: data/y_f.txt
Untracked: data/zscore_jointLCLs_m6AQTLs_susie_eQTLpruned.rds
Untracked: data/zscore_jointLCLs_random.rds
Untracked: explore_udi.R
Untracked: output/fit.k10.rds
Untracked: output/fit.nn.pbmc.purified.rds
Untracked: output/fit.nn.rds
Untracked: output/fit.nn.s.001.rds
Untracked: output/fit.nn.s.01.rds
Untracked: output/fit.nn.s.1.rds
Untracked: output/fit.nn.s.10.rds
Untracked: output/fit.snn.s.001.rds
Untracked: output/fit.snn.s.01.nninit.rds
Untracked: output/fit.snn.s.01.rds
Untracked: output/fit.varbvs.RDS
Untracked: output/fit2.nn.pbmc.purified.rds
Untracked: output/glmnet.fit.RDS
Untracked: output/snn07.txt
Untracked: output/snn34.txt
Untracked: output/test.bv.txt
Untracked: output/test.gamma.txt
Untracked: output/test.hyp.txt
Untracked: output/test.log.txt
Untracked: output/test.param.txt
Untracked: output/test2.bv.txt
Untracked: output/test2.gamma.txt
Untracked: output/test2.hyp.txt
Untracked: output/test2.log.txt
Untracked: output/test2.param.txt
Untracked: output/test3.bv.txt
Untracked: output/test3.gamma.txt
Untracked: output/test3.hyp.txt
Untracked: output/test3.log.txt
Untracked: output/test3.param.txt
Untracked: output/test4.bv.txt
Untracked: output/test4.gamma.txt
Untracked: output/test4.hyp.txt
Untracked: output/test4.log.txt
Untracked: output/test4.param.txt
Untracked: output/test5.bv.txt
Untracked: output/test5.gamma.txt
Untracked: output/test5.hyp.txt
Untracked: output/test5.log.txt
Untracked: output/test5.param.txt
Unstaged changes:
Modified: .gitignore
Modified: analysis/flashier_log1p.Rmd
Modified: analysis/flashier_sla_text.Rmd
Modified: analysis/logistic_z_scores.Rmd
Modified: analysis/mr_ash_pen.Rmd
Modified: analysis/nmu_em.Rmd
Modified: analysis/susie_flash.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/power_nneg.Rmd) and HTML
(docs/power_nneg.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5d10e53 | Matthew Stephens | 2025-03-09 | workflowr::wflow_publish("power_nneg.Rmd") |
| html | be86679 | Matthew Stephens | 2025-03-09 | Build site. |
| Rmd | 5f92e99 | Matthew Stephens | 2025-03-09 | workflowr::wflow_publish("analysis/power_nneg.Rmd") |
Introduction
I want to implement a version of the power method for symmetric nmf with \(L1\) penalty.
The idea is that given symmetric matrix \(S\) and approximation \(V D V'\) we update the \(k\)th column of \(V\) by \(V_k = [(S-UD_UU')V_k-\lambda]_+\) where \(U\) is the matrix \(V\) with the \(k\)th column removed and \(D_U\) is the matrix \(D\) with the \(k\)th row and column removed. Then we normalize \(V_k\) and set \(d_k = V_k'(S-U D_U U')V_k\).
# minimize ||S - vDv'|| + \lambda \sum abs(v) subject to v>0
# lambda is the strength of the L1 penalty
sym_nmf_update = function(S,v,d,lambda=0){
K = ncol(v)
for(k in 1:K){
U = v[,-k,drop=FALSE]
D = diag(d[-k],nrow=length(d[-k]))
newv = pmax(S %*% v[,k,drop=FALSE] - U %*% D %*% t(U) %*% v[,k,drop=FALSE] - lambda,0)
if(!all(newv==0)){
v[,k] = newv/sqrt(sum(newv^2))
} else {
v[,k] = newv
}
d[k] = t(v[,k]) %*% S %*% v[,k] - t(v[,k]) %*% U %*% D %*% t(U) %*% v[,k]
}
return(list(v=v,d=d))
}
# this is a simplified version for the rank 1 update for testing
sym_nmf_update_r1= function(S,v,d,lambda=0){
v = pmax(S %*% v - lambda,0)
v = v/sqrt(sum(v^2))
d = t(v) %*% S %*% v
return(list(v=cbind(v),d=as.vector(d)))
}
compute_sqerr = function(S,fit){
sum((S-fit$v %*% diag(fit$d,nrow=length(fit$d)) %*% t(fit$v))^2)
}Tree structured data
Simulate some data from a tree structure.
set.seed(1)
n = 40
x = cbind(c(rep(1,n),rep(0,n)), c(rep(0,n),rep(1,n)), c(rep(1,n/2),rep(0,3*n/2)), c(rep(0,n/2), rep(1,n/2), rep(0,n)), c(rep(0,n),rep(1,n/2),rep(0,n/2)), c(rep(0,3*n/2),rep(1,n/2)))
E = matrix(0.1*rexp(2*n*2*n),nrow=2*n)
E = E+t(E) #symmetric errors
A = x %*% t(x) + E
image(A)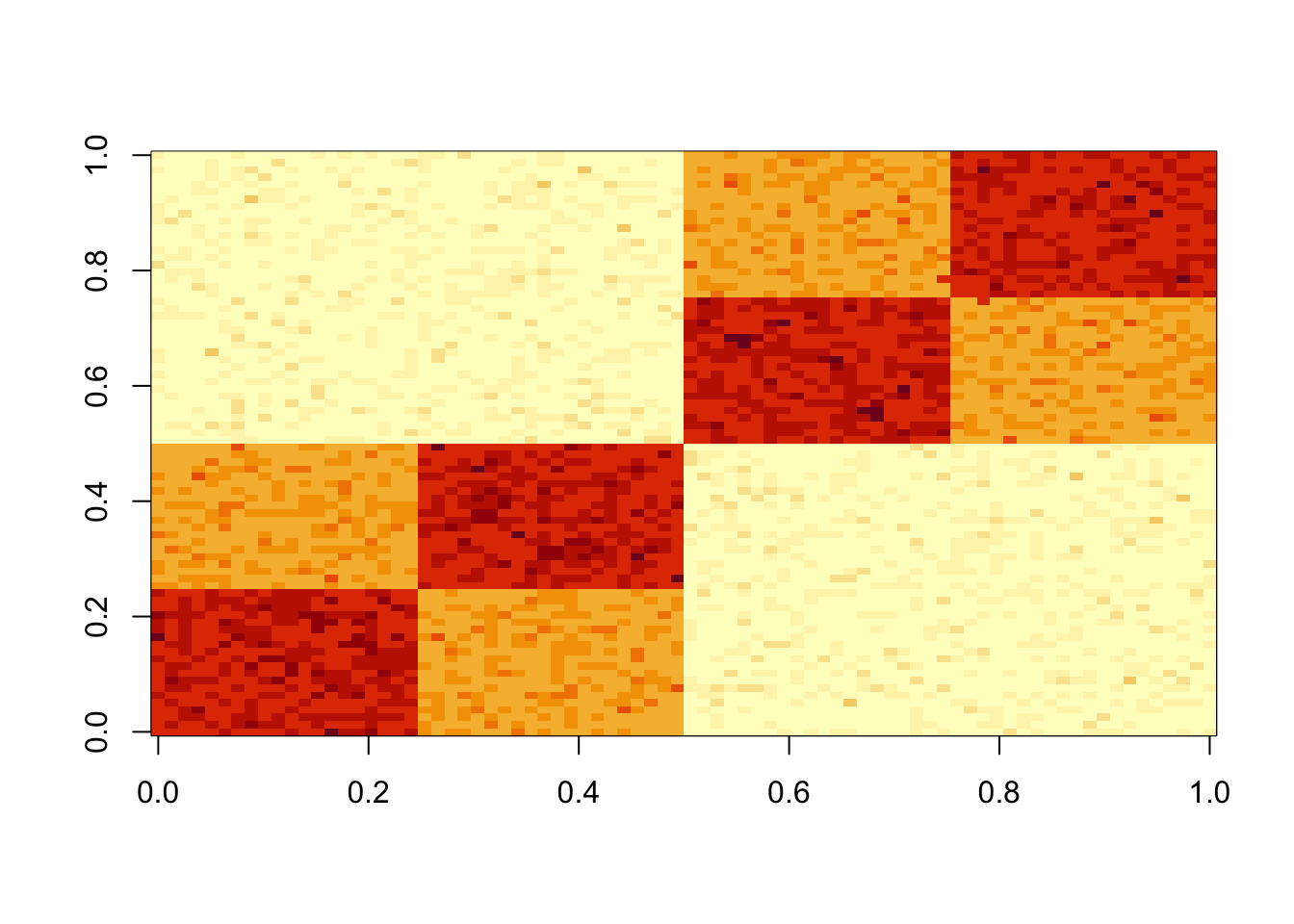
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
Rank 1 fit
Here I fit a single factor with no penalty. It finds the solution where everything is approximately equal.This is also the leading eigenvector of A, which we know to be the correct solution (because it is non-negative in this case).
set.seed(1)
K = 1
v = matrix(nrow=2*n,ncol=K)
for(k in 1:K){
v[,k] = pmax(cbind(rnorm(2*n)),0) # initialize v
v[,k] = v[,k]/sum(v[,k]^2)
}
d = rep(1,K)
fit = list(v=v,d=d)
err = rep(0,10)
err[1] = compute_sqerr(A,fit)
for(i in 2:100){
fit = sym_nmf_update(A,fit$v,fit$d)
err[i] = compute_sqerr(A,fit)
}
plot(err)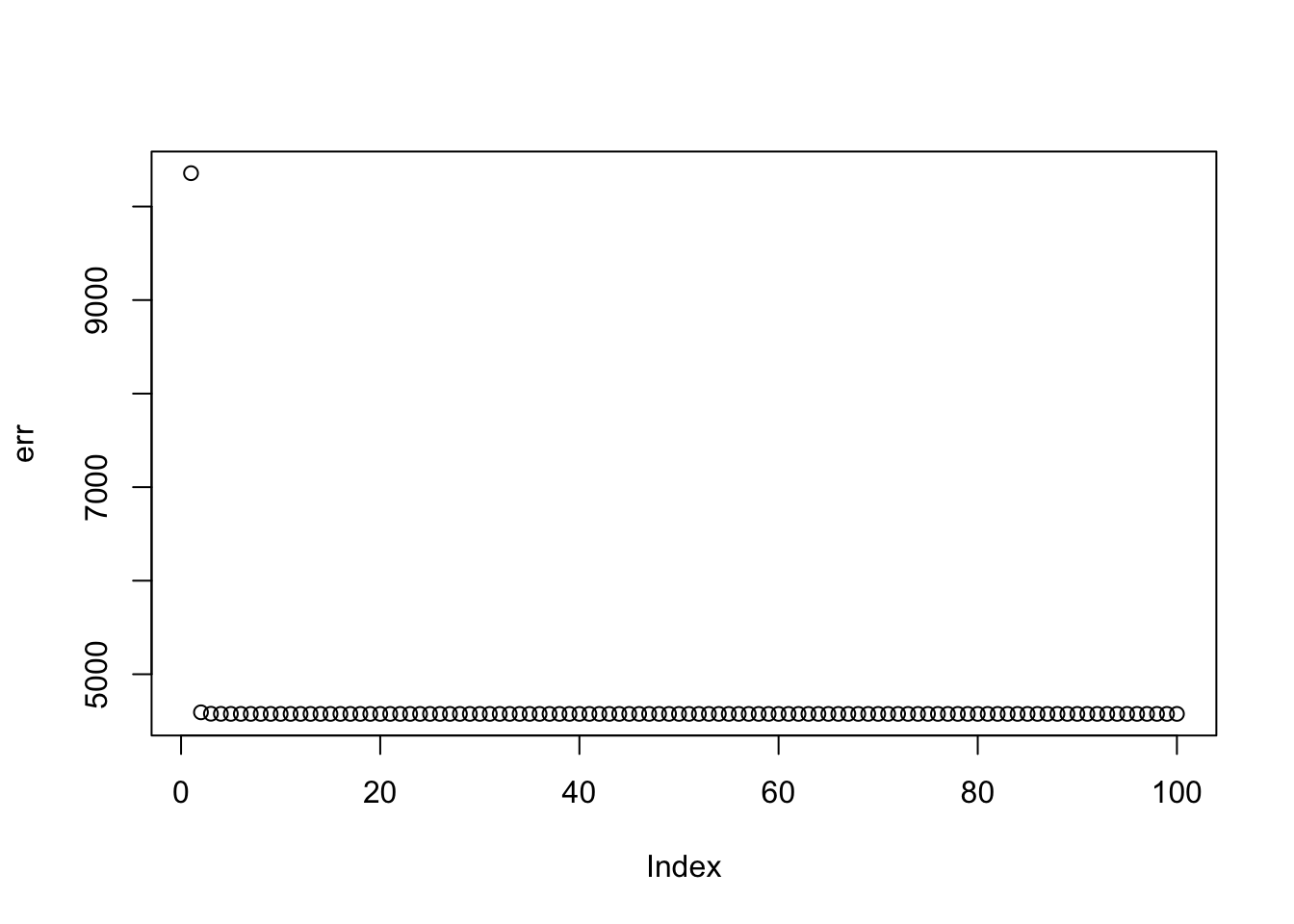
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
plot(fit$v[,1],svd(A)$v[,1])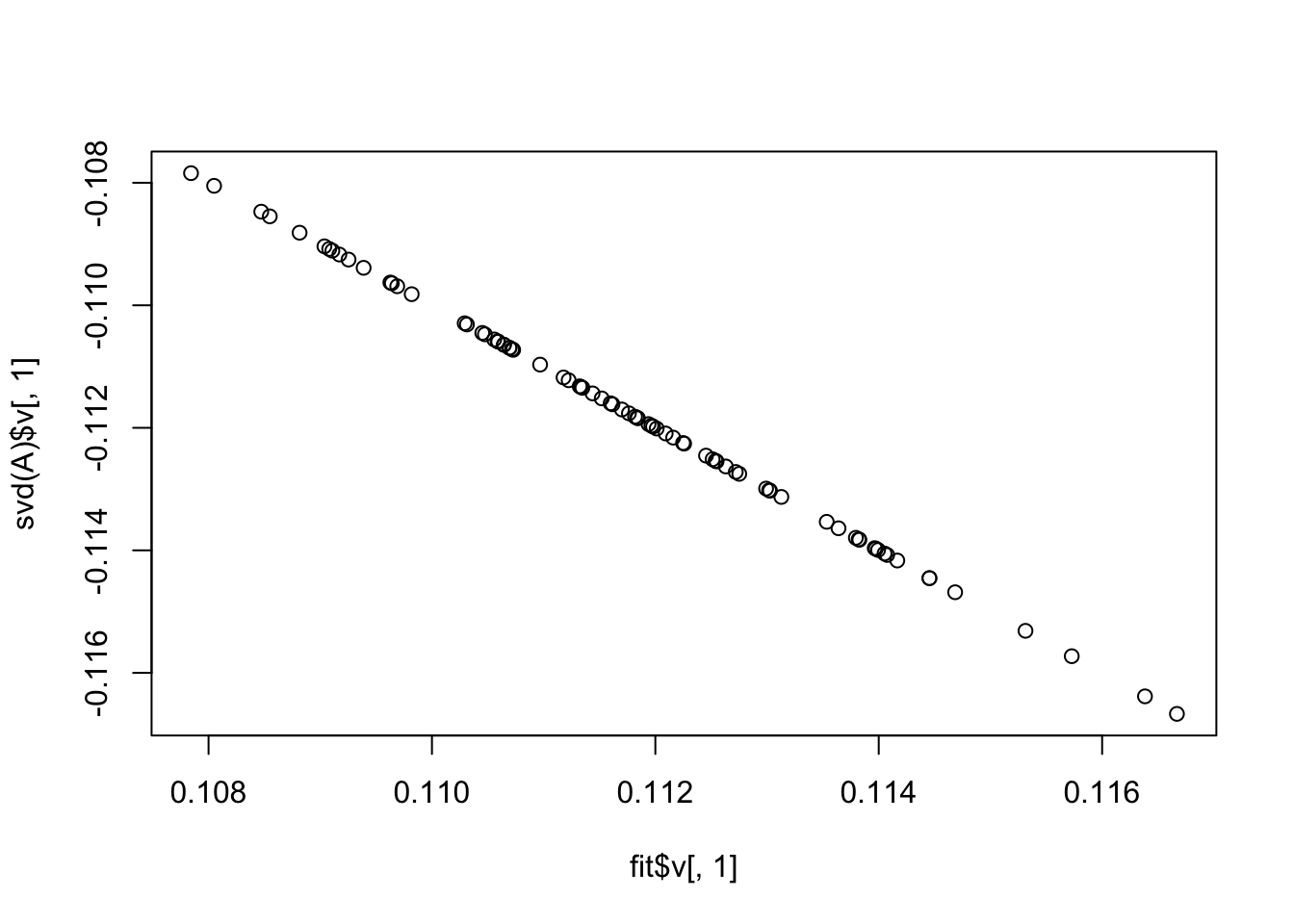
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
plot(fit$v)
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
Add penalty
Try with penalty, we are able to find a sparse solution. Note that running with penalty =2 from the start gave an error because it zeroed everything out. I had to initialize with no penalty to get it to run. It is clear that setting the penalty could be a delicate issue.
set.seed(1)
K = 1
v = matrix(nrow=2*n,ncol=K)
for(k in 1:K){
v[,k] = pmax(cbind(rnorm(2*n)),0) # initialize v
v[,k] = v[,k]/sum(v[,k]^2)
}
d = rep(1,K)
fit = list(v=v,d=d)
err = rep(0,10)
err[1] = compute_sqerr(A,fit)
for(i in 2:100){
fit = sym_nmf_update(A,fit$v,fit$d,lambda=0)
err[i] = compute_sqerr(A,fit)
}
for(i in 2:100){
fit = sym_nmf_update(A,fit$v,fit$d,lambda=2)
err[i] = compute_sqerr(A,fit)
}
plot(err)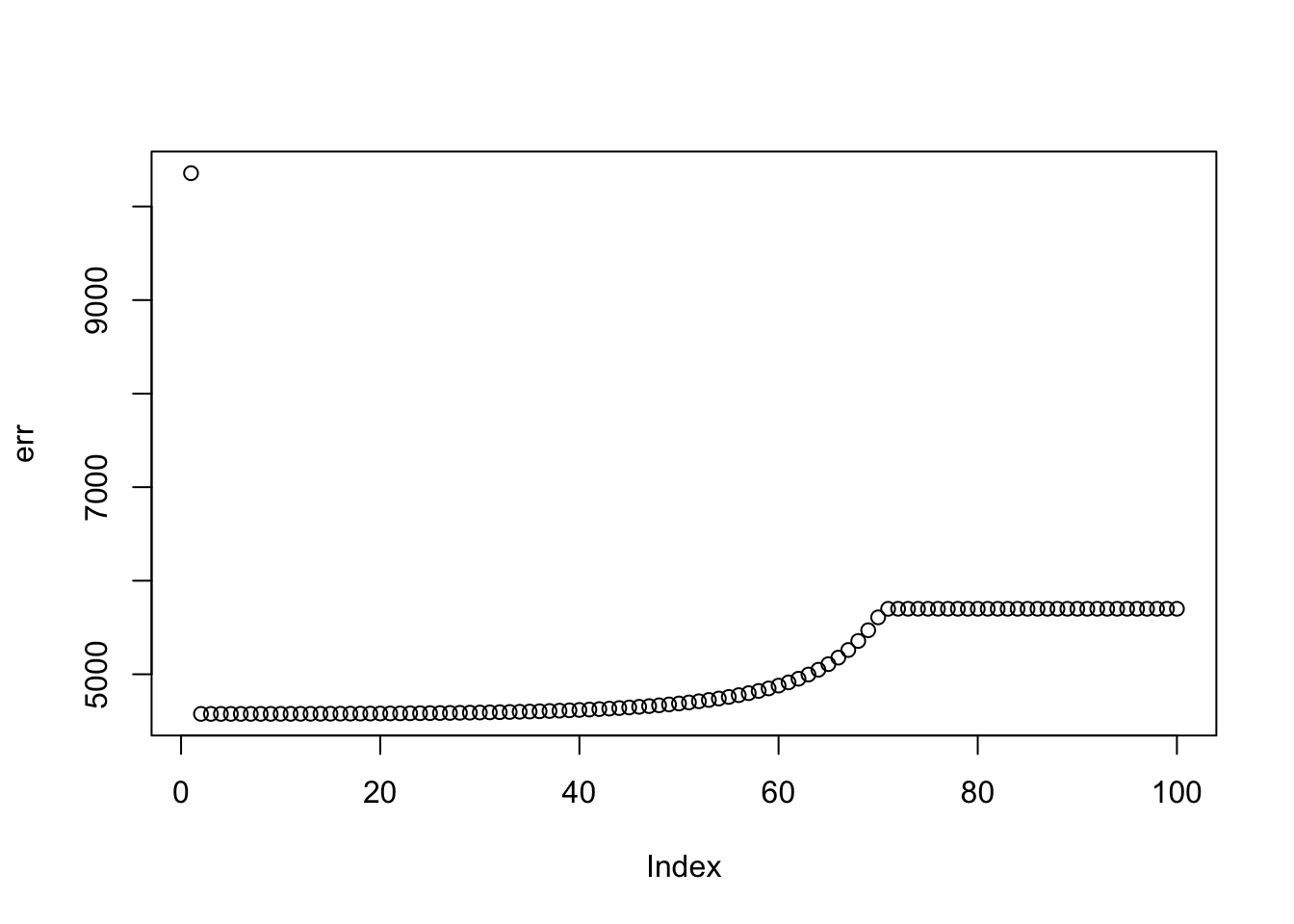
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
plot(fit$v)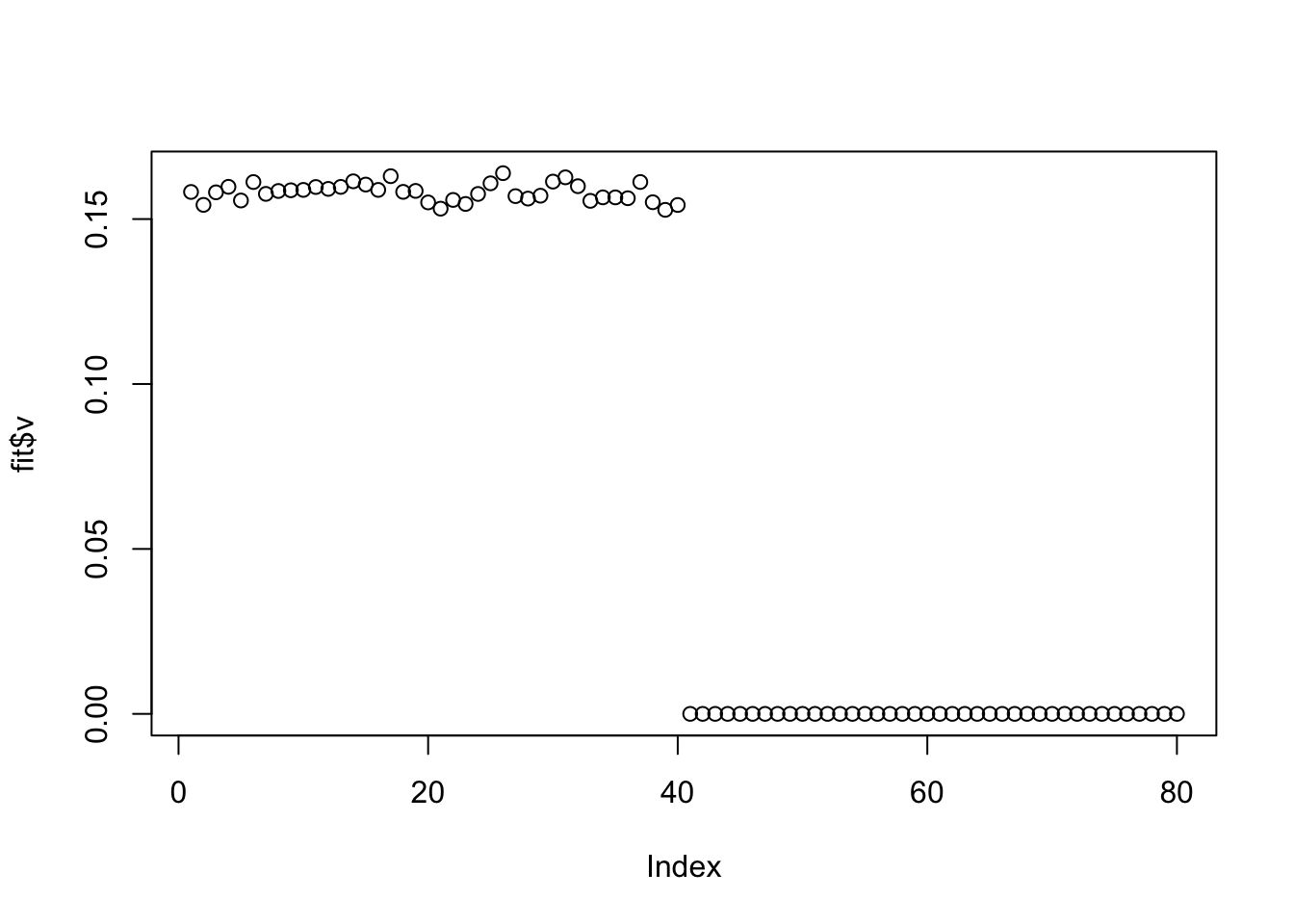
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
Run multiple factors
Here I run with no penalty and \(K=9\). It basically finds a rank 4 solution (plus 2 factors with very small weight, and 3 zero factors). Note that these solutions are already sparse (even without penalty), but not “approximately binary”. This example illustrates that, to get a tree when the truth is a tree, you need to assume more than just nonnegative and sparse. Whether it is enough to assume (approximate) binary as well is an open question.
set.seed(1)
K = 9
V = matrix(rnorm(K*2*n),ncol=K)
for(k in 1:K){
V[,k] = pmax(V[,k],0) # initialize V
V[,k] = V[,k]/sum(V[,k]^2)
}
d = rep(1,K)
fit = list(v=V,d=d)
err = rep(0,10)
err[1] = sum((A-fit$v %*% diag(fit$d) %*% t(fit$v))^2)
for(i in 2:10){
fit = sym_nmf_update(A,fit$v,fit$d)
err[i] = sum((A-fit$v %*% diag(fit$d) %*% t(fit$v))^2)
}
plot(err)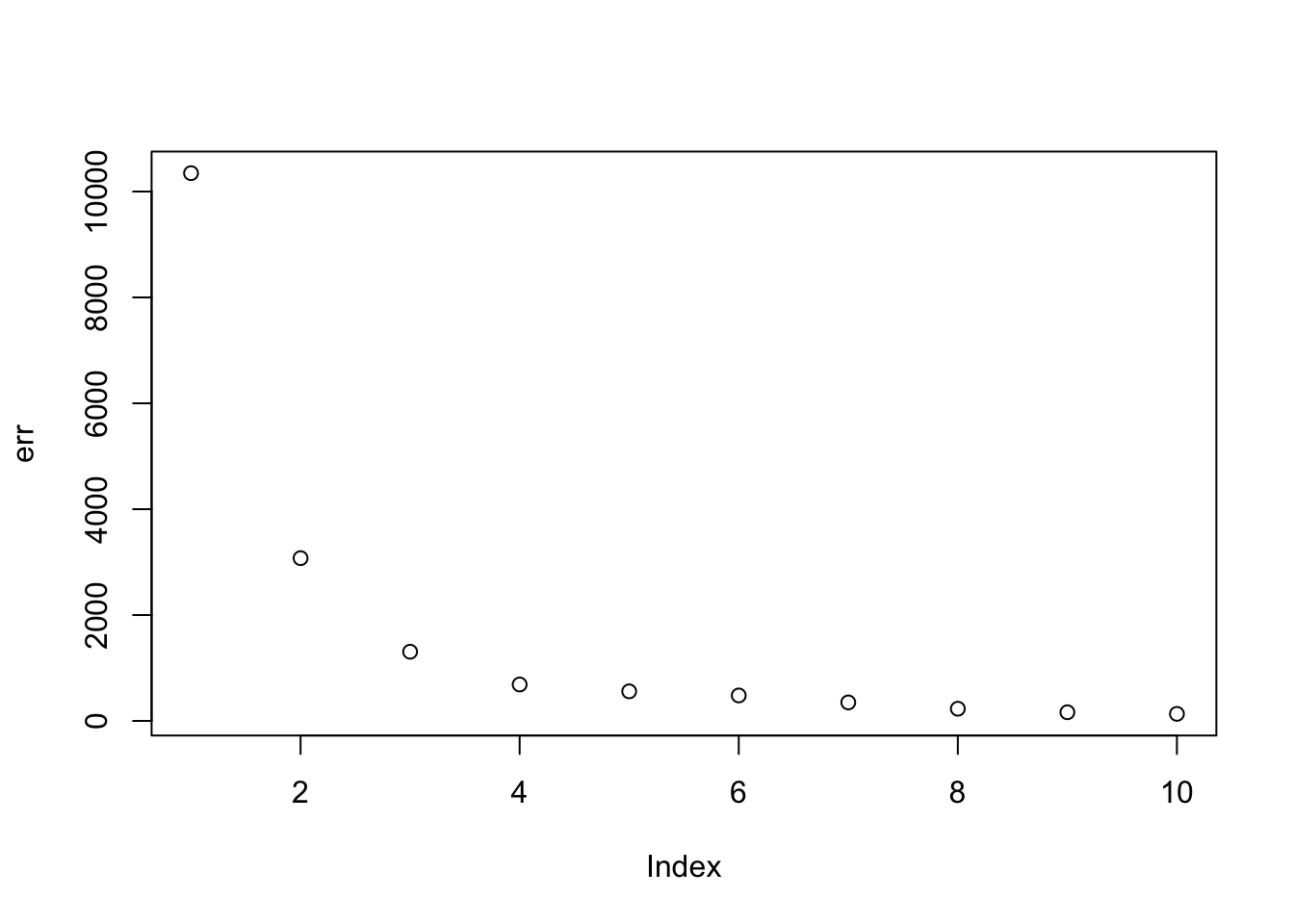
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
par(mfcol=c(3,3),mai=rep(0.1,4))
for(i in 1:9){plot(fit$v[,i],main=paste0(fit$d[i]))}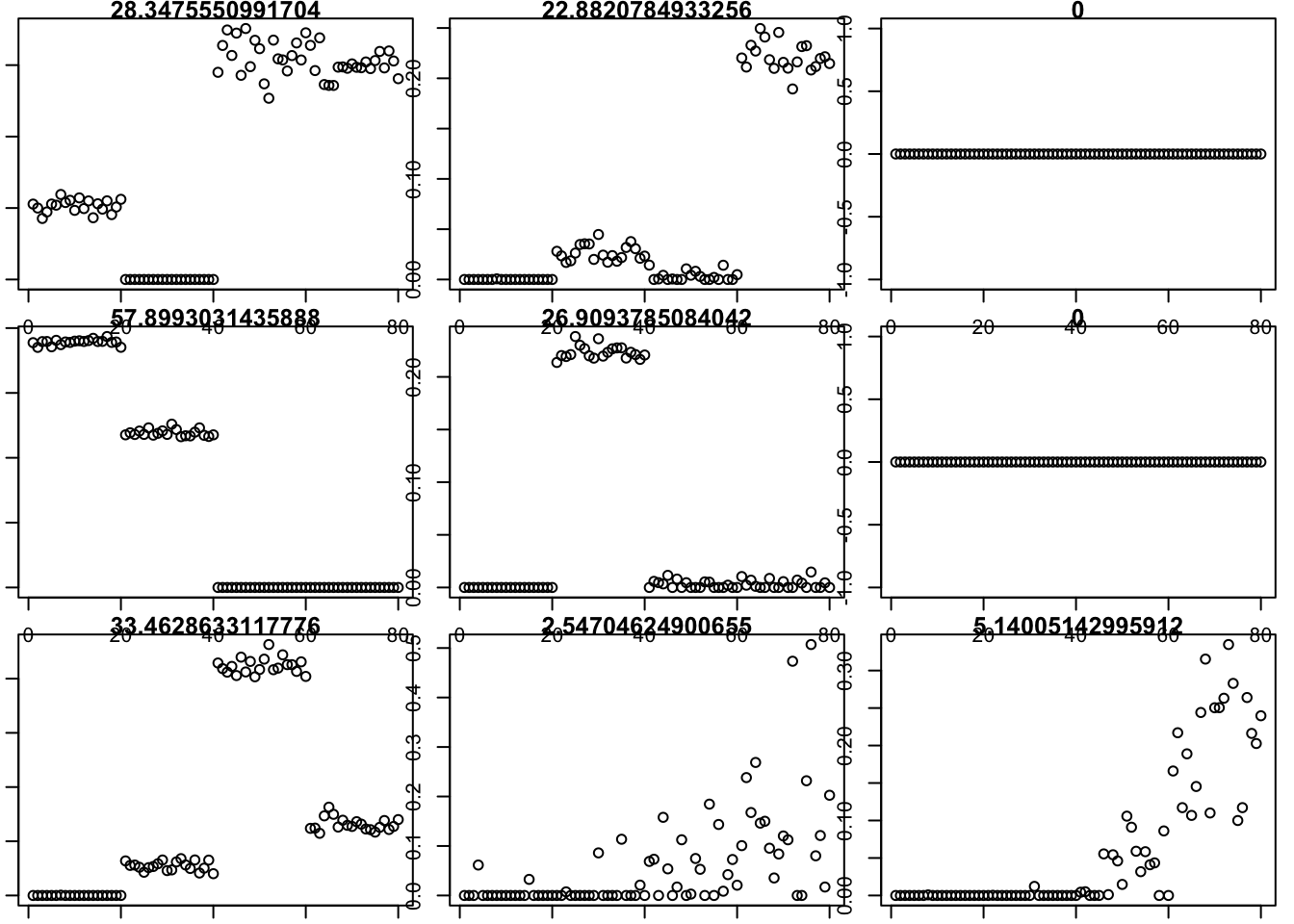
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
This shows that the solution being found has smaller error than the true value, so in that sense things seem to be working as desired.
par(mfcol=c(1,1),mai=c(0.4,0.4,0.4,0.4))
fitted = fit$v %*% diag(fit$d) %*% t(fit$v)
plot(as.vector(fitted),as.vector(A))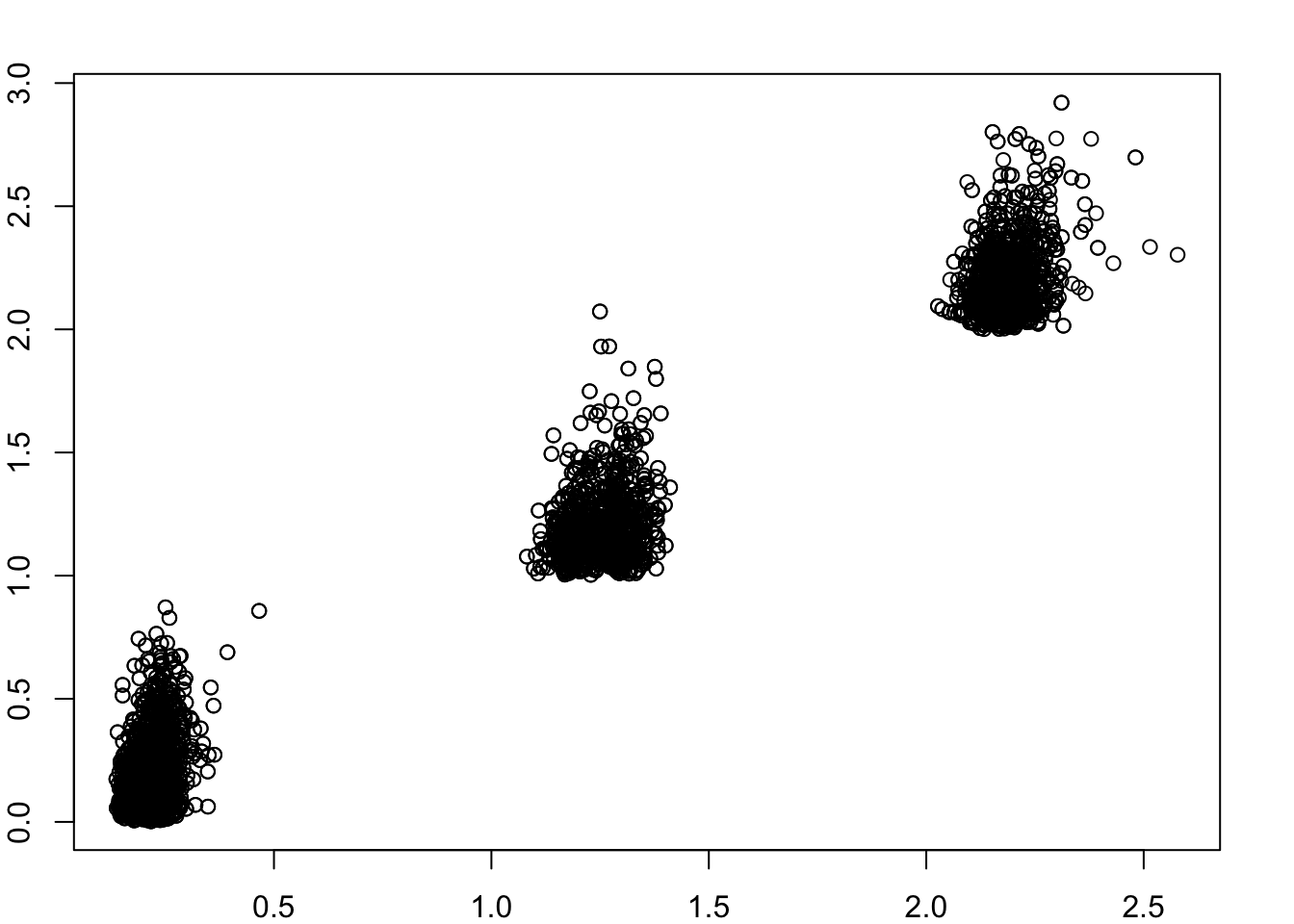
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
plot(as.vector(x %*% t(x)),as.vector(A))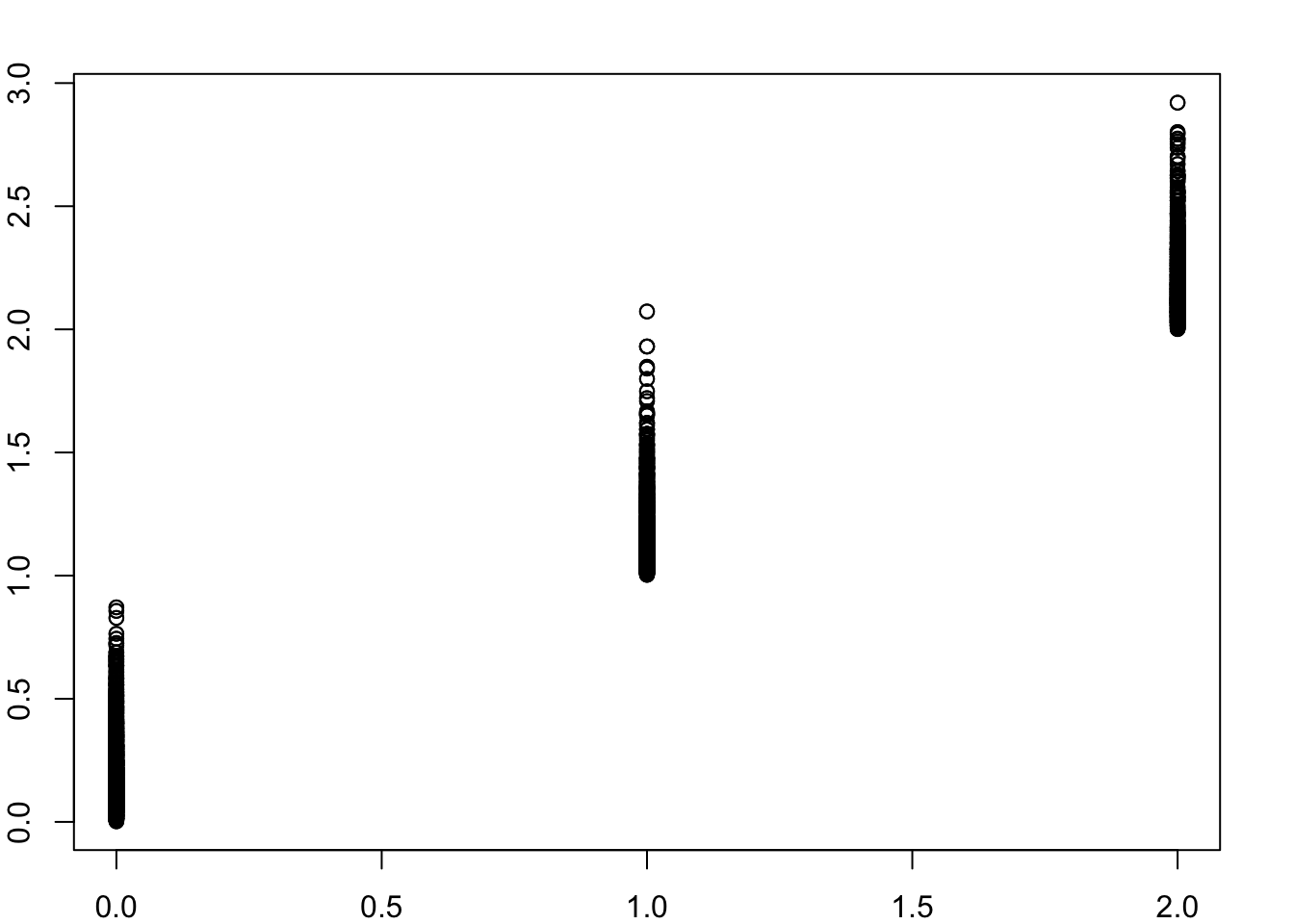
| Version | Author | Date |
|---|---|---|
| be86679 | Matthew Stephens | 2025-03-09 |
compute_sqerr(A,fit)[1] 133.0117compute_sqerr(A,fit=list(v=x,d=rep(1,6)))[1] 388.104
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Sequoia 15.3.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] vctrs_0.6.5 cli_3.6.3 knitr_1.49 rlang_1.1.5
[5] xfun_0.50 stringi_1.8.4 promises_1.3.2 jsonlite_1.8.9
[9] workflowr_1.7.1 glue_1.8.0 rprojroot_2.0.4 git2r_0.35.0
[13] htmltools_0.5.8.1 httpuv_1.6.15 sass_0.4.9 rmarkdown_2.29
[17] evaluate_1.0.3 jquerylib_0.1.4 tibble_3.2.1 fastmap_1.2.0
[21] yaml_2.3.10 lifecycle_1.0.4 whisker_0.4.1 stringr_1.5.1
[25] compiler_4.4.2 fs_1.6.5 Rcpp_1.0.14 pkgconfig_2.0.3
[29] rstudioapi_0.17.1 later_1.4.1 digest_0.6.37 R6_2.5.1
[33] pillar_1.10.1 magrittr_2.0.3 bslib_0.9.0 tools_4.4.2
[37] cachem_1.1.0