Run ebpmf pbmc3k with nonnegative LF while also updating f0
DongyueXie
2023-02-09
Last updated: 2023-02-09
Checks: 7 0
Knit directory: gsmash/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220606) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ab8513a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/movielens.Rmd
Untracked: analysis/profiled_obj_for_b_splitting.Rmd
Untracked: code/poisson_STM/structure_plot.R
Untracked: data/ml-latest-small/
Untracked: output/droplet_iteration_results/
Untracked: output/ebpmf_pbmc3k_vga3_glmpca_init.rds
Untracked: output/pbmc3k_iteration_results/
Untracked: output/pbmc_no_constraint.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/pbmc3k_nonnegLF_updatef0.Rmd) and HTML
(docs/pbmc3k_nonnegLF_updatef0.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ab8513a | DongyueXie | 2023-02-09 | wflow_publish("analysis/pbmc3k_nonnegLF_updatef0.Rmd") |
| html | 3e1fad0 | DongyueXie | 2023-02-09 | Build site. |
| Rmd | 5225a9d | DongyueXie | 2023-02-09 | wflow_publish("analysis/pbmc3k_nonnegLF_updatef0.Rmd") |
Introduction
Recall the model is
\[y_{ij}\sim Poisson(l_{i0}f_{j0}\exp(\sum_k l_{ik}f_{jk}+\epsilon_{ij}).\]
I compare the fit between fix \(f_{j0} = y_{+j}/\sqrt{y_{++}}\), and update \(f_{j0}\).
ALL other parameters are the same.
When estimating \(f_{j0}\), I got an error saying:
running initial flash fit
Running iterations...
iter 10, elbo=-18401566.4360083, K=8
iter 20, elbo=-17763539.8435548, K=8
iter 30, elbo=-17393989.0758334, K=8
Error in (function (f, p, ..., hessian = FALSE, typsize = rep(1, length(p)), :
missing value in parameter
Calls: ebpmf_log ... parametric_workhorse -> mle_parametric -> do.call -> <Anonymous>
In addition: Warning message:
In handle_standard_errors(x, s) :
Nonpositive SEs have been replaced by small positive SEs.
Execution halted
My general impression is that the point exponential prior
implementation in ebnm is less stable, and can yield less
desirable results, but much faster, when
comparing to unimodal nonnegative prior.
Since I only got results from first 30 iterations, I’ll compare the 30th iterations results.
source('code/poisson_STM/plot_factors_general.R')
source('code/poisson_STM/structure_plot.R')
library(fastTopics)
library(Matrix)
library(stm)
data(pbmc_facs)fix f0
fit = readRDS(paste('output/pbmc3k_iteration_results/ebpmf_pbmc3k_nonnegLF_pe_vga3_iter',30,'.rds',sep=''))
plt = plot.factors.general(fit$fit_flash$L.pm,pbmc_facs$samples$subpop,title=paste('pbmc3k nonneg L F iteration',30))
print(plt)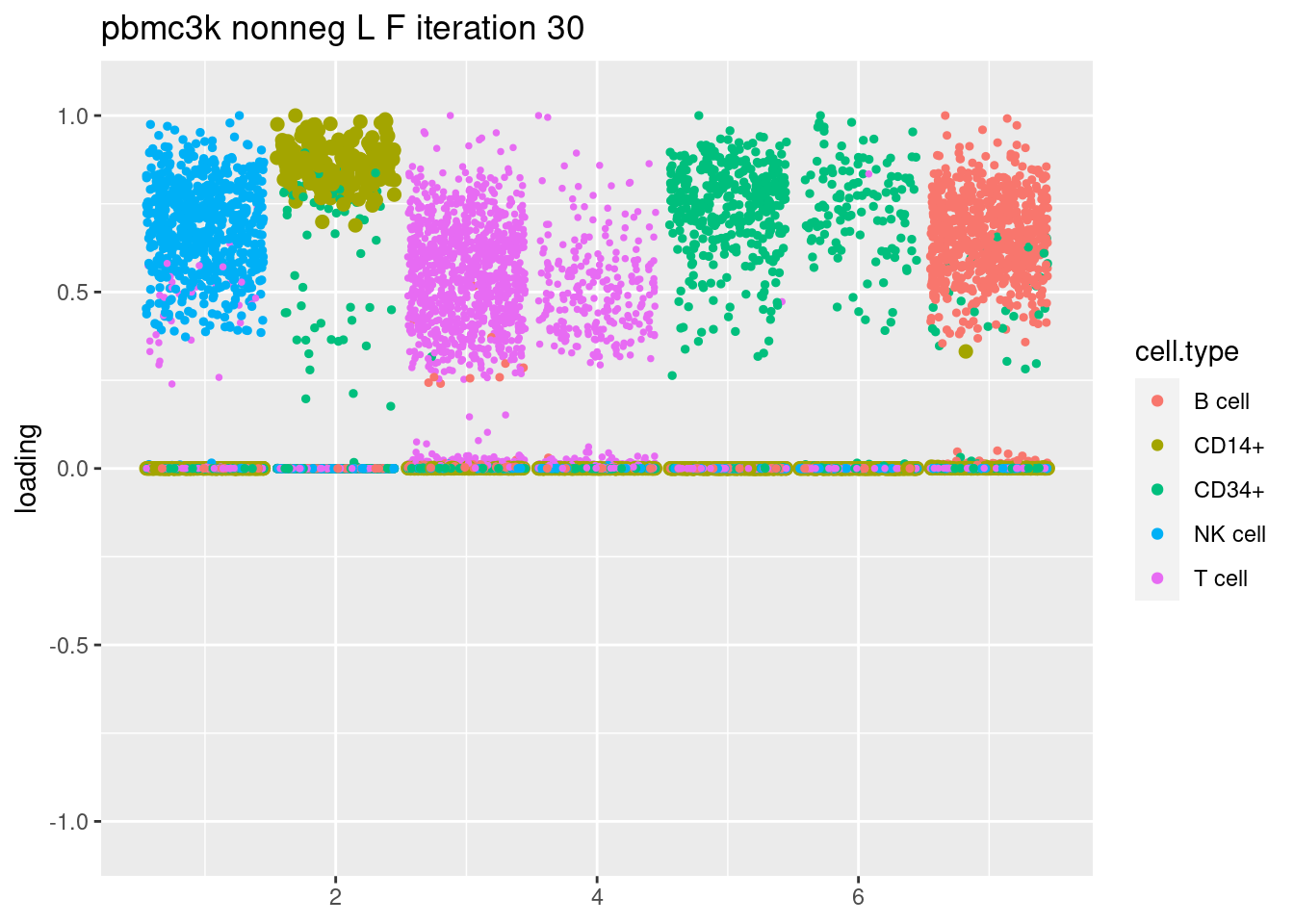
| Version | Author | Date |
|---|---|---|
| 3e1fad0 | DongyueXie | 2023-02-09 |
fit$elbo[1] -19765312structure_plot_general(fit$fit_flash$L.pm,fit$fit_flash$F.pm,pbmc_facs$samples$subpop)Running tsne on 417 x 7 matrix.Read the 417 x 7 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.07 seconds (sparsity = 0.873557)!
Learning embedding...
Iteration 50: error is 42.997166 (50 iterations in 0.04 seconds)
Iteration 100: error is 41.868908 (50 iterations in 0.03 seconds)
Iteration 150: error is 41.868062 (50 iterations in 0.03 seconds)
Iteration 200: error is 41.868149 (50 iterations in 0.04 seconds)
Iteration 250: error is 41.868091 (50 iterations in 0.03 seconds)
Iteration 300: error is 0.090940 (50 iterations in 0.03 seconds)
Iteration 350: error is 0.090530 (50 iterations in 0.03 seconds)
Iteration 400: error is 0.090535 (50 iterations in 0.03 seconds)
Iteration 450: error is 0.090537 (50 iterations in 0.03 seconds)
Iteration 500: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 550: error is 0.090537 (50 iterations in 0.03 seconds)
Iteration 600: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 650: error is 0.090537 (50 iterations in 0.03 seconds)
Iteration 700: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 750: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 800: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 850: error is 0.090537 (50 iterations in 0.03 seconds)
Iteration 900: error is 0.090535 (50 iterations in 0.03 seconds)
Iteration 950: error is 0.090538 (50 iterations in 0.03 seconds)
Iteration 1000: error is 0.090538 (50 iterations in 0.03 seconds)
Fitting performed in 0.61 seconds.Running tsne on 91 x 7 matrix.Read the 91 x 7 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 29.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.00 seconds (sparsity = 0.987320)!
Learning embedding...
Iteration 50: error is 53.493352 (50 iterations in 0.01 seconds)
Iteration 100: error is 48.356461 (50 iterations in 0.00 seconds)
Iteration 150: error is 49.518330 (50 iterations in 0.00 seconds)
Iteration 200: error is 48.106554 (50 iterations in 0.00 seconds)
Iteration 250: error is 47.422229 (50 iterations in 0.00 seconds)
Iteration 300: error is 1.112187 (50 iterations in 0.00 seconds)
Iteration 350: error is 0.697176 (50 iterations in 0.00 seconds)
Iteration 400: error is 0.690709 (50 iterations in 0.00 seconds)
Iteration 450: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 500: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 550: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 600: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 650: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 700: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 750: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 800: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 850: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 900: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 950: error is 0.690712 (50 iterations in 0.00 seconds)
Iteration 1000: error is 0.690712 (50 iterations in 0.00 seconds)
Fitting performed in 0.06 seconds.Running tsne on 358 x 7 matrix.Read the 358 x 7 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.06 seconds (sparsity = 0.951437)!
Learning embedding...
Iteration 50: error is 44.073232 (50 iterations in 0.03 seconds)
Iteration 100: error is 41.466782 (50 iterations in 0.03 seconds)
Iteration 150: error is 41.448750 (50 iterations in 0.03 seconds)
Iteration 200: error is 41.448801 (50 iterations in 0.03 seconds)
Iteration 250: error is 41.448774 (50 iterations in 0.03 seconds)
Iteration 300: error is 0.073591 (50 iterations in 0.03 seconds)
Iteration 350: error is 0.072700 (50 iterations in 0.02 seconds)
Iteration 400: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 450: error is 0.072698 (50 iterations in 0.03 seconds)
Iteration 500: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 550: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 600: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 650: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 700: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 750: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 800: error is 0.072699 (50 iterations in 0.03 seconds)
Iteration 850: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 900: error is 0.072698 (50 iterations in 0.02 seconds)
Iteration 950: error is 0.072699 (50 iterations in 0.02 seconds)
Iteration 1000: error is 0.072699 (50 iterations in 0.02 seconds)
Fitting performed in 0.49 seconds.Running tsne on 359 x 7 matrix.Read the 359 x 7 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.05 seconds (sparsity = 0.952631)!
Learning embedding...
Iteration 50: error is 41.095007 (50 iterations in 0.03 seconds)
Iteration 100: error is 40.915822 (50 iterations in 0.02 seconds)
Iteration 150: error is 40.915712 (50 iterations in 0.02 seconds)
Iteration 200: error is 40.915706 (50 iterations in 0.02 seconds)
Iteration 250: error is 40.915701 (50 iterations in 0.02 seconds)
Iteration 300: error is 0.074192 (50 iterations in 0.02 seconds)
Iteration 350: error is 0.074106 (50 iterations in 0.02 seconds)
Iteration 400: error is 0.074102 (50 iterations in 0.02 seconds)
Iteration 450: error is 0.074105 (50 iterations in 0.02 seconds)
Iteration 500: error is 0.074103 (50 iterations in 0.03 seconds)
Iteration 550: error is 0.074103 (50 iterations in 0.02 seconds)
Iteration 600: error is 0.074103 (50 iterations in 0.02 seconds)
Iteration 650: error is 0.074102 (50 iterations in 0.02 seconds)
Iteration 700: error is 0.074103 (50 iterations in 0.02 seconds)
Iteration 750: error is 0.074103 (50 iterations in 0.03 seconds)
Iteration 800: error is 0.074102 (50 iterations in 0.02 seconds)
Iteration 850: error is 0.074103 (50 iterations in 0.02 seconds)
Iteration 900: error is 0.074103 (50 iterations in 0.03 seconds)
Iteration 950: error is 0.074103 (50 iterations in 0.03 seconds)
Iteration 1000: error is 0.074103 (50 iterations in 0.02 seconds)
Fitting performed in 0.48 seconds.Running tsne on 775 x 7 matrix.Read the 775 x 7 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.12 seconds (sparsity = 0.498091)!
Learning embedding...
Iteration 50: error is 45.482085 (50 iterations in 0.07 seconds)
Iteration 100: error is 41.615579 (50 iterations in 0.06 seconds)
Iteration 150: error is 41.017730 (50 iterations in 0.07 seconds)
Iteration 200: error is 40.805126 (50 iterations in 0.06 seconds)
Iteration 250: error is 40.708478 (50 iterations in 0.06 seconds)
Iteration 300: error is 0.183027 (50 iterations in 0.06 seconds)
Iteration 350: error is 0.125610 (50 iterations in 0.06 seconds)
Iteration 400: error is 0.116738 (50 iterations in 0.06 seconds)
Iteration 450: error is 0.114335 (50 iterations in 0.07 seconds)
Iteration 500: error is 0.113508 (50 iterations in 0.06 seconds)
Iteration 550: error is 0.113136 (50 iterations in 0.06 seconds)
Iteration 600: error is 0.112980 (50 iterations in 0.07 seconds)
Iteration 650: error is 0.112861 (50 iterations in 0.06 seconds)
Iteration 700: error is 0.112738 (50 iterations in 0.06 seconds)
Iteration 750: error is 0.112710 (50 iterations in 0.06 seconds)
Iteration 800: error is 0.112659 (50 iterations in 0.06 seconds)
Iteration 850: error is 0.112640 (50 iterations in 0.06 seconds)
Iteration 900: error is 0.112656 (50 iterations in 0.06 seconds)
Iteration 950: error is 0.112646 (50 iterations in 0.06 seconds)
Iteration 1000: error is 0.112634 (50 iterations in 0.06 seconds)
Fitting performed in 1.26 seconds.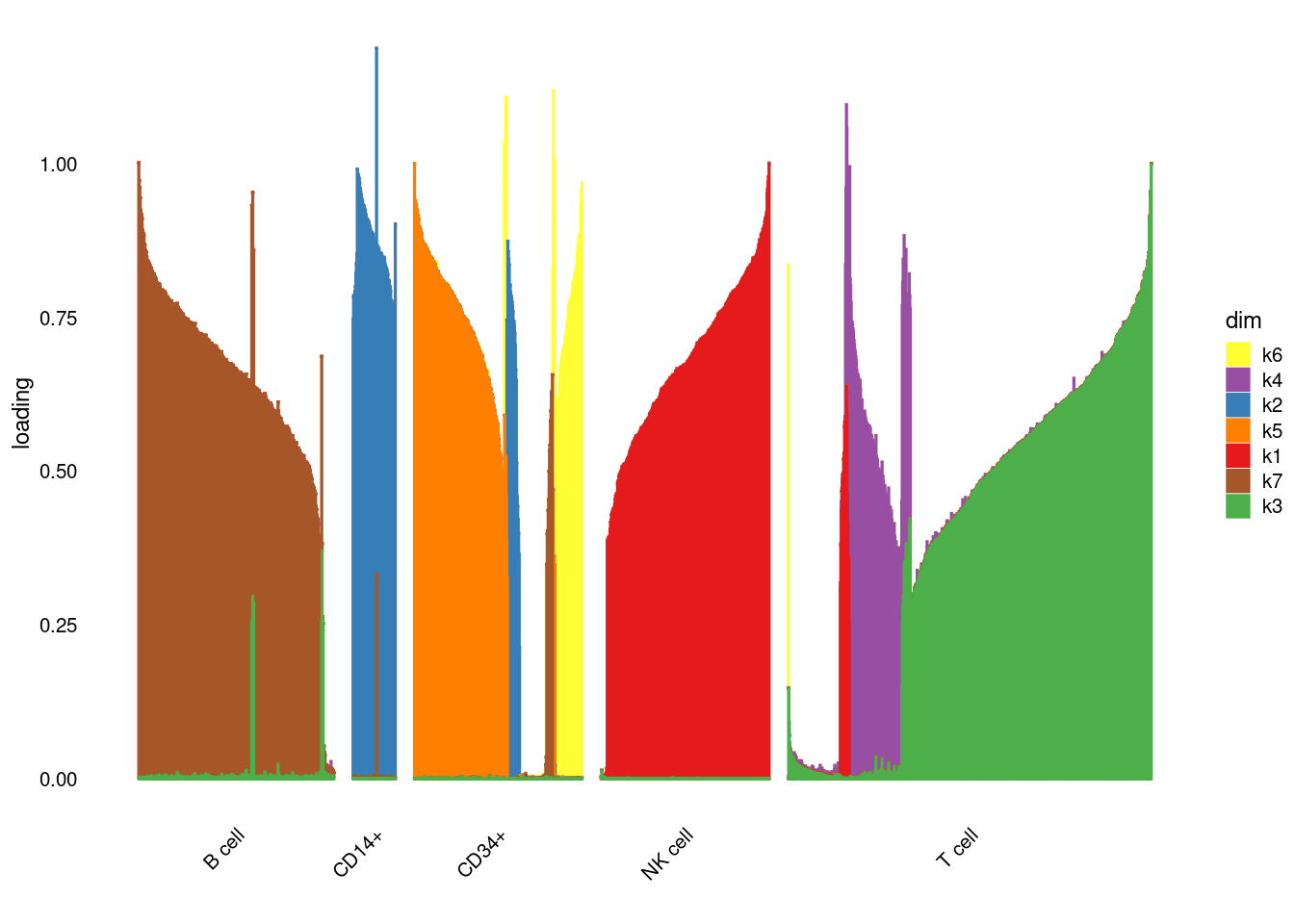
f00 = colSums(pbmc_facs$counts)/sqrt(sum(pbmc_facs$counts))update f0
fit = readRDS(paste('output/pbmc3k_iteration_results/ebpmf_pbmc3k_nonnegLF_pe_vga3_est_f0_iter30.rds'))
plt = plot.factors.general(fit$fit_flash$L.pm,pbmc_facs$samples$subpop,title=paste('pbmc3k update f0 nonneg L F iteration',30))
print(plt)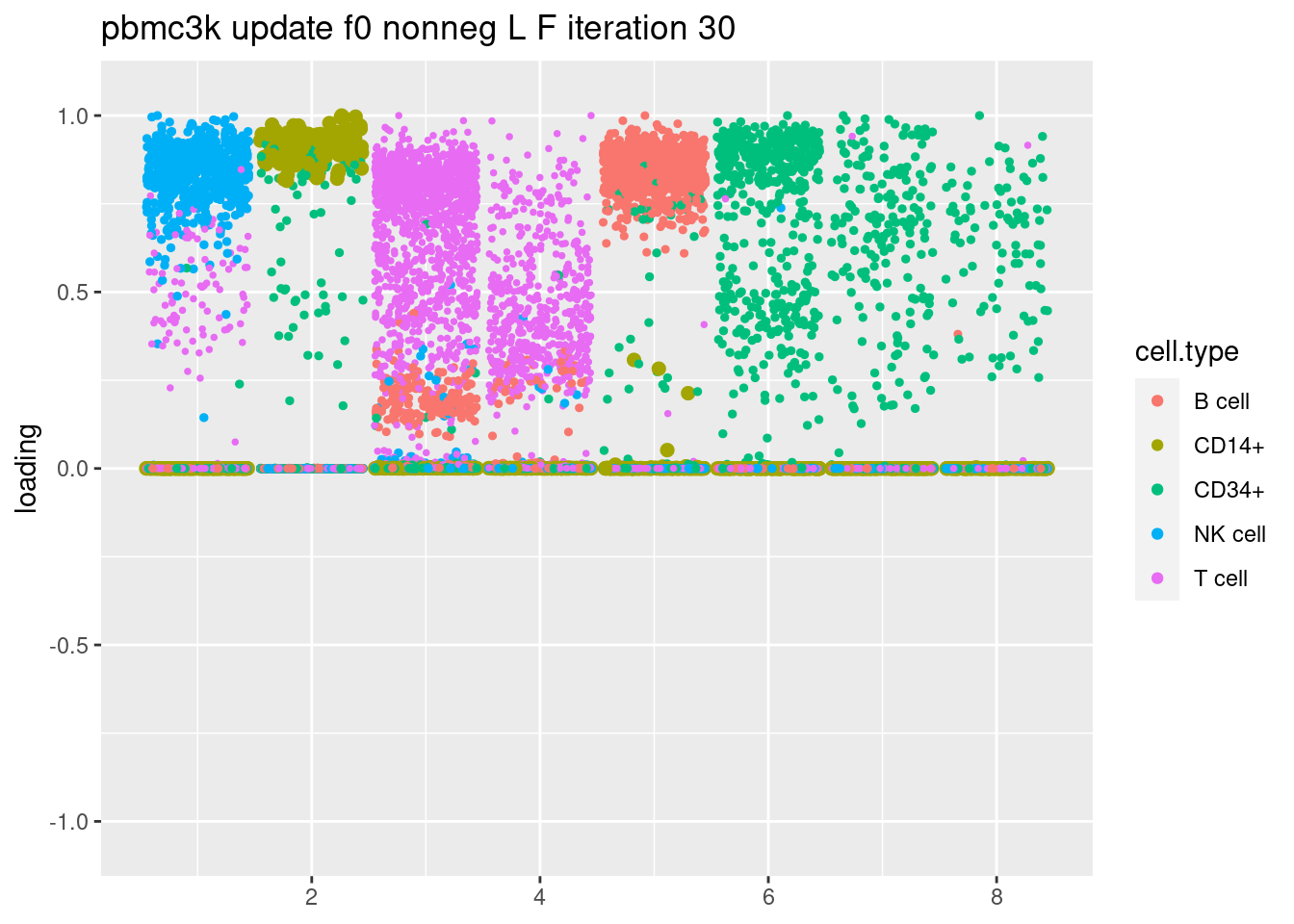
| Version | Author | Date |
|---|---|---|
| 3e1fad0 | DongyueXie | 2023-02-09 |
fit$elbo[1] -19580914structure_plot_general(fit$fit_flash$L.pm,fit$fit_flash$F.pm,pbmc_facs$samples$subpop)Running tsne on 417 x 8 matrix.Read the 417 x 8 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.07 seconds (sparsity = 0.875306)!
Learning embedding...
Iteration 50: error is 45.701851 (50 iterations in 0.04 seconds)
Iteration 100: error is 41.959043 (50 iterations in 0.03 seconds)
Iteration 150: error is 41.904270 (50 iterations in 0.03 seconds)
Iteration 200: error is 41.903176 (50 iterations in 0.03 seconds)
Iteration 250: error is 41.903098 (50 iterations in 0.04 seconds)
Iteration 300: error is 0.092093 (50 iterations in 0.03 seconds)
Iteration 350: error is 0.087865 (50 iterations in 0.03 seconds)
Iteration 400: error is 0.087711 (50 iterations in 0.03 seconds)
Iteration 450: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 500: error is 0.087729 (50 iterations in 0.03 seconds)
Iteration 550: error is 0.087731 (50 iterations in 0.03 seconds)
Iteration 600: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 650: error is 0.087731 (50 iterations in 0.03 seconds)
Iteration 700: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 750: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 800: error is 0.087731 (50 iterations in 0.03 seconds)
Iteration 850: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 900: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 950: error is 0.087732 (50 iterations in 0.03 seconds)
Iteration 1000: error is 0.087732 (50 iterations in 0.03 seconds)
Fitting performed in 0.58 seconds.Running tsne on 91 x 8 matrix.Read the 91 x 8 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 29.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.00 seconds (sparsity = 0.986837)!
Learning embedding...
Iteration 50: error is 49.735471 (50 iterations in 0.00 seconds)
Iteration 100: error is 48.569182 (50 iterations in 0.00 seconds)
Iteration 150: error is 50.621830 (50 iterations in 0.00 seconds)
Iteration 200: error is 50.788143 (50 iterations in 0.00 seconds)
Iteration 250: error is 52.077153 (50 iterations in 0.00 seconds)
Iteration 300: error is 1.641253 (50 iterations in 0.00 seconds)
Iteration 350: error is 0.800636 (50 iterations in 0.00 seconds)
Iteration 400: error is 0.796971 (50 iterations in 0.00 seconds)
Iteration 450: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 500: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 550: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 600: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 650: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 700: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 750: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 800: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 850: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 900: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 950: error is 0.796977 (50 iterations in 0.00 seconds)
Iteration 1000: error is 0.796977 (50 iterations in 0.00 seconds)
Fitting performed in 0.06 seconds.Running tsne on 358 x 8 matrix.Read the 358 x 8 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.05 seconds (sparsity = 0.950953)!
Learning embedding...
Iteration 50: error is 43.493234 (50 iterations in 0.03 seconds)
Iteration 100: error is 43.493234 (50 iterations in 0.03 seconds)
Iteration 150: error is 43.493234 (50 iterations in 0.03 seconds)
Iteration 200: error is 43.493234 (50 iterations in 0.03 seconds)
Iteration 250: error is 43.493234 (50 iterations in 0.03 seconds)
Iteration 300: error is 0.164546 (50 iterations in 0.03 seconds)
Iteration 350: error is 0.160839 (50 iterations in 0.02 seconds)
Iteration 400: error is 0.159392 (50 iterations in 0.03 seconds)
Iteration 450: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 500: error is 0.159391 (50 iterations in 0.02 seconds)
Iteration 550: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 600: error is 0.159391 (50 iterations in 0.02 seconds)
Iteration 650: error is 0.159391 (50 iterations in 0.02 seconds)
Iteration 700: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 750: error is 0.159391 (50 iterations in 0.02 seconds)
Iteration 800: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 850: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 900: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 950: error is 0.159392 (50 iterations in 0.02 seconds)
Iteration 1000: error is 0.159392 (50 iterations in 0.02 seconds)
Fitting performed in 0.51 seconds.Running tsne on 359 x 8 matrix.Read the 359 x 8 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.05 seconds (sparsity = 0.952615)!
Learning embedding...
Iteration 50: error is 43.105018 (50 iterations in 0.03 seconds)
Iteration 100: error is 41.222909 (50 iterations in 0.03 seconds)
Iteration 150: error is 41.222379 (50 iterations in 0.02 seconds)
Iteration 200: error is 41.222379 (50 iterations in 0.02 seconds)
Iteration 250: error is 41.222380 (50 iterations in 0.02 seconds)
Iteration 300: error is 0.090568 (50 iterations in 0.02 seconds)
Iteration 350: error is 0.090346 (50 iterations in 0.02 seconds)
Iteration 400: error is 0.090346 (50 iterations in 0.02 seconds)
Iteration 450: error is 0.090345 (50 iterations in 0.02 seconds)
Iteration 500: error is 0.090345 (50 iterations in 0.03 seconds)
Iteration 550: error is 0.090346 (50 iterations in 0.03 seconds)
Iteration 600: error is 0.090346 (50 iterations in 0.04 seconds)
Iteration 650: error is 0.090346 (50 iterations in 0.04 seconds)
Iteration 700: error is 0.090346 (50 iterations in 0.02 seconds)
Iteration 750: error is 0.090346 (50 iterations in 0.03 seconds)
Iteration 800: error is 0.090345 (50 iterations in 0.03 seconds)
Iteration 850: error is 0.090346 (50 iterations in 0.03 seconds)
Iteration 900: error is 0.090346 (50 iterations in 0.03 seconds)
Iteration 950: error is 0.090345 (50 iterations in 0.03 seconds)
Iteration 1000: error is 0.090345 (50 iterations in 0.04 seconds)
Fitting performed in 0.58 seconds.Running tsne on 775 x 8 matrix.Read the 775 x 8 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 1, perplexity = 100.000000, and theta = 0.100000
Computing input similarities...
Building tree...
Done in 0.15 seconds (sparsity = 0.500172)!
Learning embedding...
Iteration 50: error is 46.009739 (50 iterations in 0.07 seconds)
Iteration 100: error is 43.831645 (50 iterations in 0.06 seconds)
Iteration 150: error is 43.565172 (50 iterations in 0.09 seconds)
Iteration 200: error is 43.487247 (50 iterations in 0.09 seconds)
Iteration 250: error is 43.457697 (50 iterations in 0.06 seconds)
Iteration 300: error is 0.240611 (50 iterations in 0.07 seconds)
Iteration 350: error is 0.186438 (50 iterations in 0.09 seconds)
Iteration 400: error is 0.178554 (50 iterations in 0.09 seconds)
Iteration 450: error is 0.176758 (50 iterations in 0.07 seconds)
Iteration 500: error is 0.176225 (50 iterations in 0.06 seconds)
Iteration 550: error is 0.176036 (50 iterations in 0.06 seconds)
Iteration 600: error is 0.175941 (50 iterations in 0.06 seconds)
Iteration 650: error is 0.175917 (50 iterations in 0.06 seconds)
Iteration 700: error is 0.175878 (50 iterations in 0.06 seconds)
Iteration 750: error is 0.175807 (50 iterations in 0.07 seconds)
Iteration 800: error is 0.175806 (50 iterations in 0.06 seconds)
Iteration 850: error is 0.175784 (50 iterations in 0.08 seconds)
Iteration 900: error is 0.175774 (50 iterations in 0.08 seconds)
Iteration 950: error is 0.175774 (50 iterations in 0.06 seconds)
Iteration 1000: error is 0.175777 (50 iterations in 0.07 seconds)
Fitting performed in 1.44 seconds.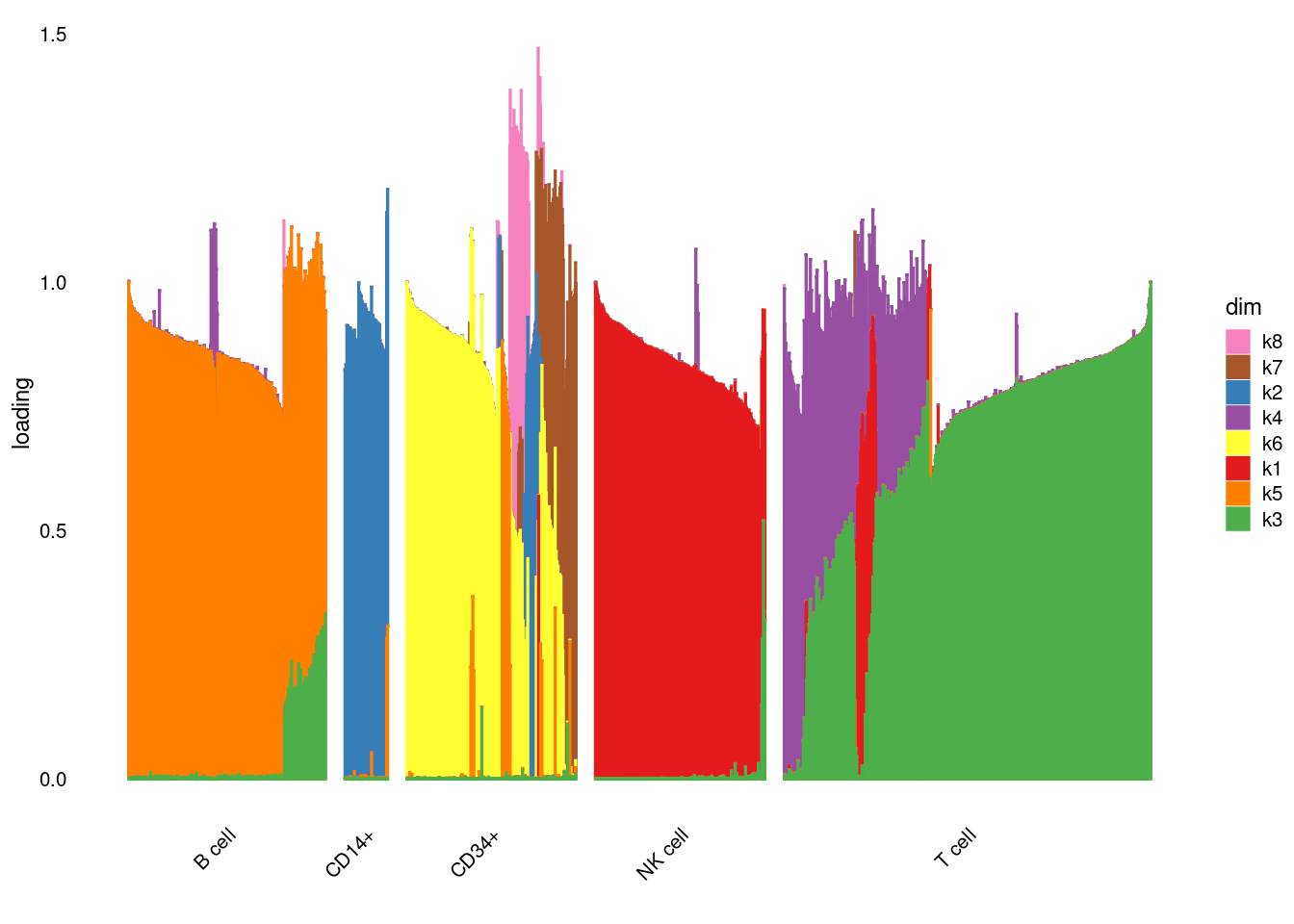
plot(f00,fit$f0,xlab='init f0',ylab='fitted f0',main='fixed vs updated')
abline(a=0,b=1,lty=2)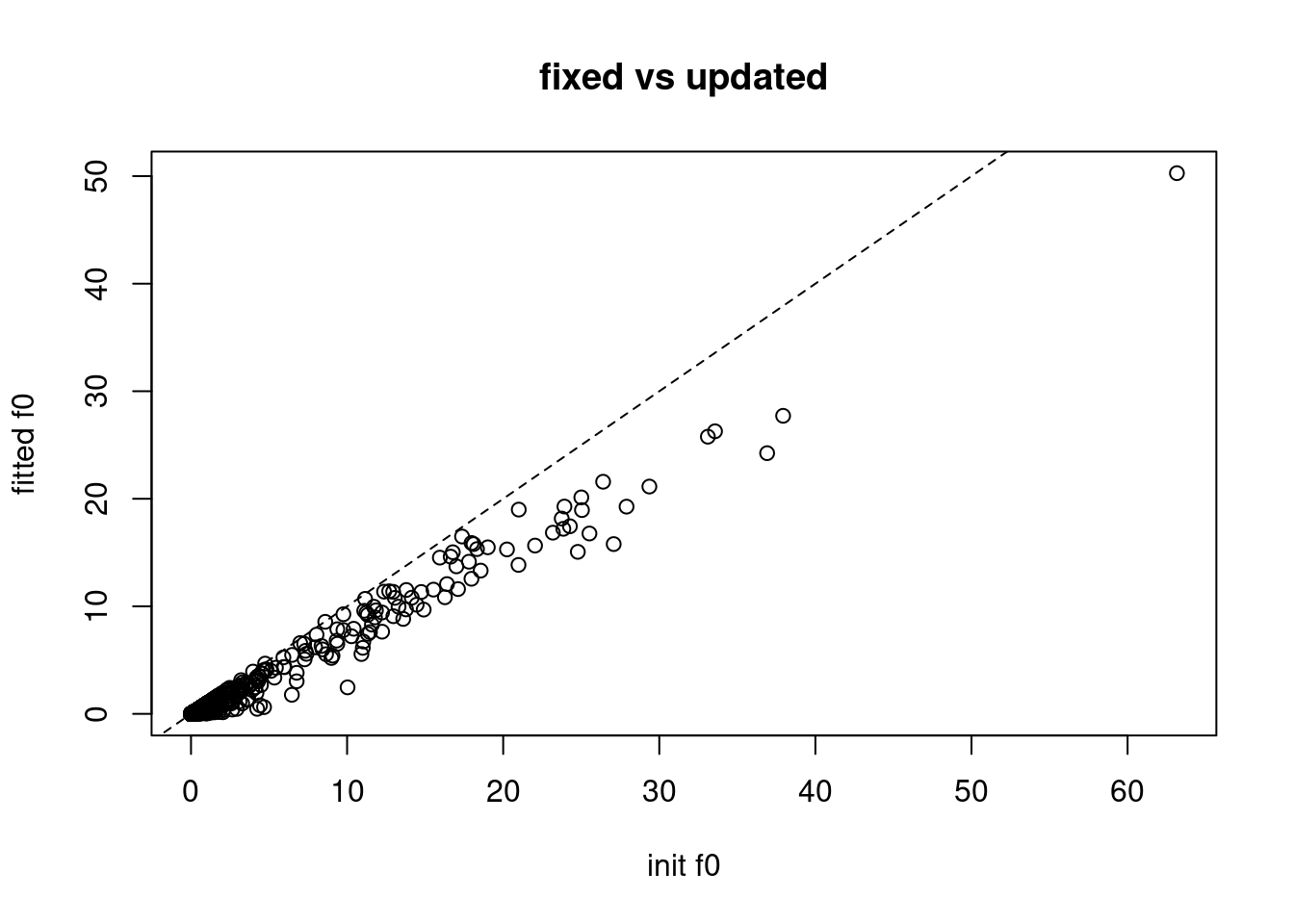
sessionInfo()R version 4.2.2 Patched (2022-11-10 r83330)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] stm_1.3.5 Matrix_1.5-3 fastTopics_0.6-142 ggplot2_3.4.0
[5] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] Rtsne_0.16 ebpm_0.0.1.3 colorspace_2.0-3
[4] smashr_1.3-6 ellipsis_0.3.2 mr.ash_0.1-87
[7] rprojroot_2.0.3 fs_1.5.2 rstudioapi_0.14
[10] farver_2.1.1 MatrixModels_0.5-1 ggrepel_0.9.2
[13] fansi_1.0.3 codetools_0.2-19 splines_4.2.2
[16] cachem_1.0.6 knitr_1.41 jsonlite_1.8.4
[19] nloptr_2.0.3 mcmc_0.9-7 ashr_2.2-54
[22] smashrgen_1.1.5 uwot_0.1.14 compiler_4.2.2
[25] httr_1.4.4 RcppZiggurat_0.1.6 fastmap_1.1.0
[28] lazyeval_0.2.2 cli_3.4.1 later_1.3.0
[31] htmltools_0.5.4 quantreg_5.94 prettyunits_1.1.1
[34] tools_4.2.2 coda_0.19-4 gtable_0.3.1
[37] glue_1.6.2 reshape2_1.4.4 dplyr_1.0.10
[40] Rcpp_1.0.9 softImpute_1.4-1 jquerylib_0.1.4
[43] vctrs_0.5.1 iterators_1.0.14 wavethresh_4.7.2
[46] xfun_0.35 stringr_1.5.0 ps_1.7.2
[49] trust_0.1-8 lifecycle_1.0.3 irlba_2.3.5.1
[52] NNLM_0.4.4 getPass_0.2-2 MASS_7.3-58.2
[55] scales_1.2.1 hms_1.1.2 promises_1.2.0.1
[58] parallel_4.2.2 SparseM_1.81 yaml_2.3.6
[61] pbapply_1.6-0 sass_0.4.4 stringi_1.7.8
[64] SQUAREM_2021.1 highr_0.9 deconvolveR_1.2-1
[67] foreach_1.5.2 caTools_1.18.2 truncnorm_1.0-8
[70] shape_1.4.6 horseshoe_0.2.0 rlang_1.0.6
[73] pkgconfig_2.0.3 matrixStats_0.63.0 bitops_1.0-7
[76] ebnm_1.0-11 evaluate_0.19 lattice_0.20-45
[79] invgamma_1.1 purrr_0.3.5 labeling_0.4.2
[82] htmlwidgets_1.6.0 Rfast_2.0.6 cowplot_1.1.1
[85] processx_3.8.0 tidyselect_1.2.0 plyr_1.8.8
[88] magrittr_2.0.3 R6_2.5.1 generics_0.1.3
[91] pillar_1.8.1 whisker_0.4.1 withr_2.5.0
[94] survival_3.5-0 mixsqp_0.3-48 tibble_3.1.8
[97] crayon_1.5.2 utf8_1.2.2 plotly_4.10.1
[100] rmarkdown_2.19 progress_1.2.2 grid_4.2.2
[103] data.table_1.14.6 callr_3.7.3 git2r_0.30.1
[106] digest_0.6.31 vebpm_0.4.0 tidyr_1.2.1
[109] httpuv_1.6.7 MCMCpack_1.6-3 RcppParallel_5.1.5
[112] munsell_0.5.0 glmnet_4.1-6 viridisLite_0.4.1
[115] flashier_0.2.34 bslib_0.4.2 quadprog_1.5-8